
Welcome to the web supplement accompanying the manuscript: "Phylogenetic Portrait of the S. cerevisiae Functional Genome ". Here, you will find searchable interfaces, supplemental figures, and downloadable data, figures, and methods..
Please contact biocomp@princeton.edu with any issues.
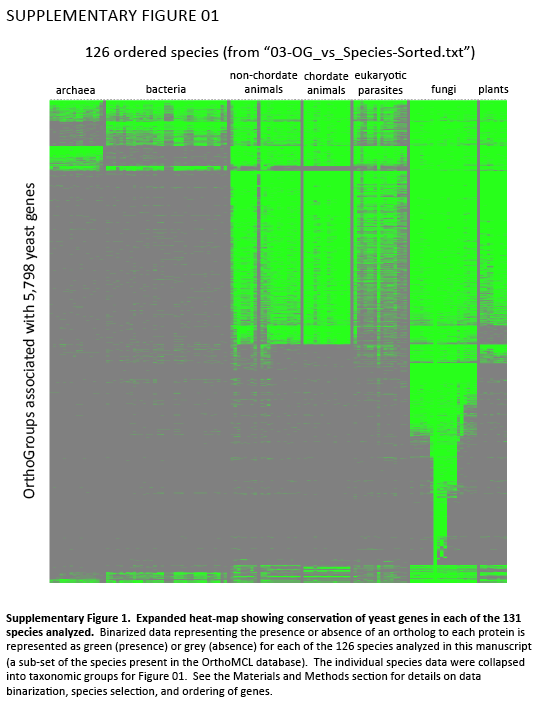
Supplementary Figure 1. Expanded heat-map showing conservation of yeast genes in each of the 131 species analyzed. Binarized data representing the presence or absence of an ortholog to each protein is represented as green (presence) or grey (absence) for each of the 126 species analyzed in this manuscript (a sub-set of the species present in the OrthoMCL database). The individual species data were collapsed into taxonomic groups for Figure 01. See the Materials and Methods section for details on data binarization, species selection, and ordering of genes.

Supplementary Figure 2. Fine-scale analysis of Minor Phylogroups. Expanded view of the Minor Phylogroups with included labels for rough phylogenetic categories to the right. An asterisk (*) indicates that only one gene is present with the identified phylogenetic pattern, and due to space limitations is not fully described in the phylogenetic categories to the right.
Supplementary Figure 3. Phylogroup-based analysis of non-annotated/non-deleted genes in S. cerevisiae. Phylogroup-based analysis of the 325 genes that have no data in Saccharomyces Genome Database (SGD) for deletion-based phenotypes. These genes fall into two categories: a) genes that have no reported deletion made, and b) genes that are present in the non-essential updated version of the haploid deletion collection from OpenBiosystems, but no primary literature has been published listing them as non-essential (and therefore these cannot be curated as non-essential). Inclusion of this gene set into this manuscript now constitutes publication of their phenotype, and thus basis for curation into SGD.
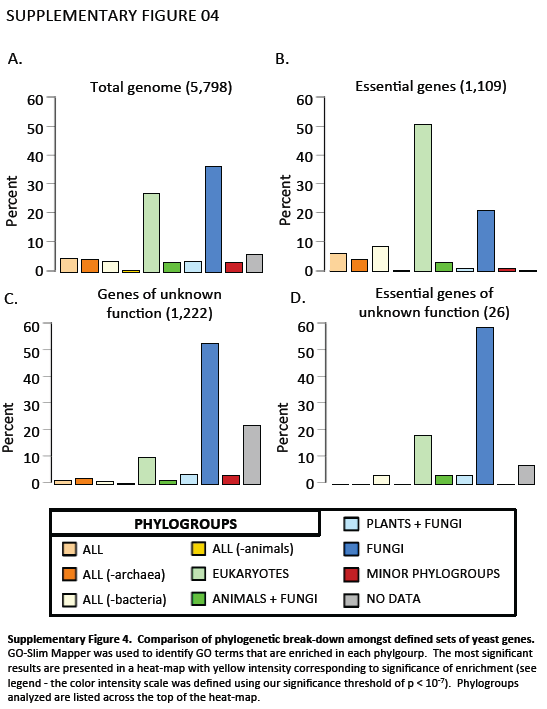
Supplementary Figure 4. Gene Ontology (GO) functional category term enrichment of phylogroups. GO-Slim Mapper was used to identify GO terms that are enriched in each phylgroup. The most significant results are presented in a heat-map with yellow intensity corresponding to significance of enrichment (see legend - the color intensity scale was defined using our significance threshold of p < 10-7). Phylogroups analyzed are listed across the top of the heat-map.
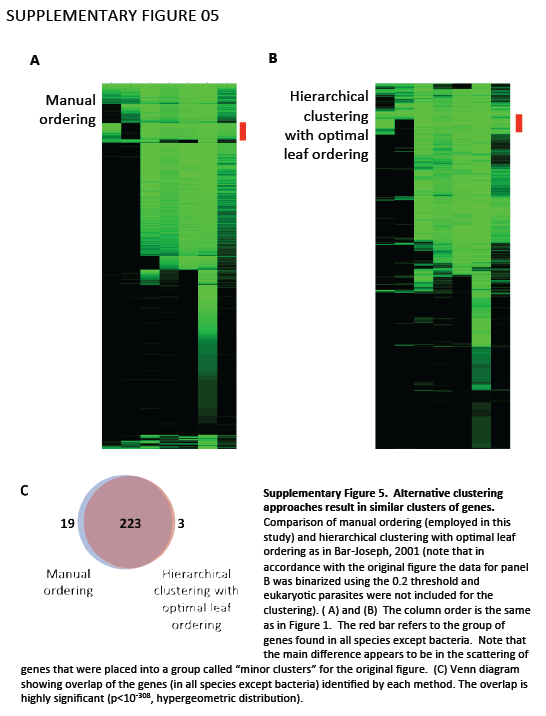
Supplementary Figure 5. Alternative clustering approaches result in similar clusters of genes. Comparison of manual ordering (employed in this study) and hierarchical clustering with optimal leaf ordering as in Bar-Joseph, 2001 (note that in accordance with the original figure the data for panel B was binarized using the 0.2 threshold and eukaryotic parasites were not included for the clustering). (A) and (B) The column order is the same as in Figure 1. The red bar refers to the group of genes found in all species except bacteria. Note that the main difference appears to be in the scattering of genes that were placed into a group called "minor clusters" for the original figure. (C) Venn diagram showing overlap of the genes (in all species except bacteria) identified by each method. The overlap is highly significant (p<10-308, hypergeometric distribution).
| Systematic Name: | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Standard Name: | |||||||||||||||
| External Links: | |||||||||||||||
| Description: | |||||||||||||||
| Conservation: |
|
||||||||||||||
| Phylogroup: |
| PhyloGroup | GO-Category | GO term | log2(fold change) | P-value (BY corrected) |
|---|---|---|---|---|
| 0 - ALL | Component | cell cortex | -2.727044607 | 0.2131435 |
| 0 - ALL | Component | cell wall | 0.093002728 | 1 |
| 0 - ALL | Component | cellular bud | -1.144712363 | 0.9232297 |
| 0 - ALL | Component | cellular component unknown | -0.9429033 | 0.02436707 |
| 0 - ALL | Component | chromosome | -1.457677953 | 0.06176952 |
| 0 - ALL | Component | cytoplasm | 0.463339758 | 0 |
| 0 - ALL | Component | cytoplasmic membrane-bounded vesicle | -1.370479511 | 1 |
| 0 - ALL | Component | cytoskeleton | -1.021329947 | 0.9322268 |
| 0 - ALL | Component | endomembrane system | -1.301437866 | 0.09455167 |
| 0 - ALL | Component | endoplasmic reticulum | -1.195066563 | 0.1168117 |
| 0 - ALL | Component | extracellular region | 1.201062474 | 0.7288225 |
| 0 - ALL | Component | Golgi apparatus | -1.583208835 | 0.3987875 |
| 0 - ALL | Component | membrane | -0.76130848 | 0.000989976 |
| 0 - ALL | Component | membrane fraction | -0.15257448 | 1 |
| 0 - ALL | Component | microtubule organizing center | -0.818837083 | 1 |
| 0 - ALL | Component | mitochondrial envelope | 0.21448299 | 1 |
| 0 - ALL | Component | mitochondrion | 1.012795837 | 3.99E-13 |
| 0 - ALL | Component | nucleolus | -1.619967385 | 0.04823153 |
| 0 - ALL | Component | nucleus | -0.503019758 | 0.004383737 |
| 0 - ALL | Component | peroxisome | 0.348619663 | 1 |
| 0 - ALL | Component | plasma membrane | -0.996583285 | 0.5517671 |
| 0 - ALL | Component | ribosome | 0.435117813 | 0.9232297 |
| 0 - ALL | Component | site of polarized growth | -0.64820609 | 1 |
| 0 - ALL | Component | vacuole | -0.107984329 | 1 |
| 0 - ALL | Function | ATPase activity | -0.474502575 | 1 |
| 0 - ALL | Function | cytoskeletal protein binding | -0.651380337 | 1 |
| 0 - ALL | Function | DNA binding | -1.461902539 | 0.06000298 |
| 0 - ALL | Function | enzyme binding | (none in phylogroup) | 1 |
| 0 - ALL | Function | enzyme regulator activity | -1.812234348 | 0.1471212 |
| 0 - ALL | Function | GTPase activity | (none in phylogroup) | 0.971396 |
| 0 - ALL | Function | helicase activity | (none in phylogroup) | 0.4077548 |
| 0 - ALL | Function | histone binding | (none in phylogroup) | 1 |
| 0 - ALL | Function | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 2.527712979 | 2.85E-08 |
| 0 - ALL | Function | hydrolase activity, acting on glycosyl bonds | 0.816398624 | 1 |
| 0 - ALL | Function | ion binding | 0.598397972 | 1 |
| 0 - ALL | Function | isomerase activity | 1.708022463 | 0.01338807 |
| 0 - ALL | Function | kinase activity | 0.297738494 | 1 |
| 0 - ALL | Function | ligase activity | 2.271451802 | 0 |
| 0 - ALL | Function | lipid binding | (none in phylogroup) | 0.3029601 |
| 0 - ALL | Function | lyase activity | 2.51951 | 6.05E-11 |
| 0 - ALL | Function | methyltransferase activity | 0.529685222 | 1 |
| 0 - ALL | Function | molecular function unknown | -1.949497202 | 2.15E-21 |
| 0 - ALL | Function | mRNA binding | 0.708022463 | 1 |
| 0 - ALL | Function | nuclease activity | 0.155974585 | 1 |
| 0 - ALL | Function | nucleic acid binding transcription factor activity | (none in phylogroup) | 0.3269333 |
| 0 - ALL | Function | nucleotidyltransferase activity | -0.127263392 | 1 |
| 0 - ALL | Function | oxidoreductase activity | 2.017350521 | 0 |
| 0 - ALL | Function | peptidase activity | -0.727044607 | 1 |
| 0 - ALL | Function | phosphatase activity | -0.228925367 | 1 |
| 0 - ALL | Function | protein binding transcription factor activity | (none in phylogroup) | 0.483015 |
| 0 - ALL | Function | protein binding, bridging | (none in phylogroup) | 1 |
| 0 - ALL | Function | protein transporter activity | (none in phylogroup) | 1 |
| 0 - ALL | Function | RNA binding | -1.48699352 | 0.000113766 |
| 0 - ALL | Function | RNA modification guide activity | (none in phylogroup) | 0.586864 |
| 0 - ALL | Function | rRNA binding | -1.168171336 | 1 |
| 0 - ALL | Function | signal transducer activity | 0.049059381 | 1 |
| 0 - ALL | Function | small conjugating protein binding | (none in phylogroup) | 1 |
| 0 - ALL | Function | structural constituent of ribosome | 0.962404998 | 0.03568229 |
| 0 - ALL | Function | structural molecule activity | 0.295699348 | 1 |
| 0 - ALL | Function | transcription factor binding | (none in phylogroup) | 1 |
| 0 - ALL | Function | transferase activity, transferring acyl groups | -0.436367446 | 1 |
| 0 - ALL | Function | transferase activity, transferring alkyl or aryl (other than methyl) groups | 2.085585257 | 0.003506821 |
| 0 - ALL | Function | transferase activity, transferring glycosyl groups | 0.727131286 | 0.9027281 |
| 0 - ALL | Function | translation factor activity, nucleic acid binding | (none in phylogroup) | 1 |
| 0 - ALL | Function | transmembrane transporter activity | -0.195359347 | 1 |
| 0 - ALL | Function | triplet codon-amino acid adaptor activity | (none in phylogroup) | 2.01E-05 |
| 0 - ALL | Function | unfolded protein binding | -0.110139214 | 1 |
| 0 - ALL | Process | amino acid transport | (none in phylogroup) | 1 |
| 0 - ALL | Process | biological process unknown | -1.119677728 | 1.93E-05 |
| 0 - ALL | Process | carbohydrate metabolic process | 1.442880394 | 7.67E-07 |
| 0 - ALL | Process | carbohydrate transport | -0.876940038 | 1 |
| 0 - ALL | Process | cell budding | -1.038403461 | 1 |
| 0 - ALL | Process | cell morphogenesis | 0.16153411 | 1 |
| 0 - ALL | Process | cell wall organization or biogenesis | -0.403799584 | 1 |
| 0 - ALL | Process | cellular amino acid metabolic process | 2.613843999 | 0 |
| 0 - ALL | Process | cellular ion homeostasis | 0.887904589 | 0.3449095 |
| 0 - ALL | Process | cellular respiration | 1.654780442 | 0.001474119 |
| 0 - ALL | Process | chromatin organization | -1.397196849 | 0.3803804 |
| 0 - ALL | Process | chromosome segregation | -1.120865621 | 1 |
| 0 - ALL | Process | cofactor metabolic process | 2.457402227 | 0 |
| 0 - ALL | Process | conjugation | -0.914414743 | 1 |
| 0 - ALL | Process | cytokinesis | -0.825409737 | 1 |
| 0 - ALL | Process | cytoplasmic translation | 1.097968981 | 0.02429649 |
| 0 - ALL | Process | cytoskeleton organization | -1.831952583 | 0.132819 |
| 0 - ALL | Process | DNA recombination | -1.828684869 | 0.5005812 |
| 0 - ALL | Process | DNA repair | -0.667147653 | 1 |
| 0 - ALL | Process | DNA replication | 0.262463019 | 1 |
| 0 - ALL | Process | endocytosis | (none in phylogroup) | 0.2085557 |
| 0 - ALL | Process | endosome transport | (none in phylogroup) | 0.5016732 |
| 0 - ALL | Process | exocytosis | (none in phylogroup) | 1 |
| 0 - ALL | Process | generation of precursor metabolites and energy | 1.46409688 | 0.000222115 |
| 0 - ALL | Process | Golgi vesicle transport | -1.144712363 | 0.9232297 |
| 0 - ALL | Process | histone modification | -0.629012524 | 1 |
| 0 - ALL | Process | invasive growth in response to glucose limitation | 0.283524634 | 1 |
| 0 - ALL | Process | ion transport | -0.662435526 | 1 |
| 0 - ALL | Process | lipid metabolic process | -0.492510524 | 1 |
| 0 - ALL | Process | lipid transport | (none in phylogroup) | 1 |
| 0 - ALL | Process | meiotic cell cycle | -2.977740679 | 0.07842359 |
| 0 - ALL | Process | membrane fusion | 0.407513352 | 1 |
| 0 - ALL | Process | membrane invagination | (none in phylogroup) | 0.03712361 |
| 0 - ALL | Process | mitochondrial translation | 1.101526801 | 0.0638186 |
| 0 - ALL | Process | mitochondrion organization | 0.670547757 | 0.1873545 |
| 0 - ALL | Process | mitotic cell cycle | -2.193797143 | 0.01473155 |
| 0 - ALL | Process | mRNA processing | -3.112828302 | 0.04256697 |
| 0 - ALL | Process | nuclear transport | -0.125438351 | 1 |
| 0 - ALL | Process | nucleobase, nucleoside and nucleotide metabolic process | 2.462135364 | 0 |
| 0 - ALL | Process | nucleobase, nucleoside, nucleotide and nucleic acid transport | 0.345452383 | 1 |
| 0 - ALL | Process | nucleus organization | (none in phylogroup) | 0.971396 |
| 0 - ALL | Process | oligosaccharide metabolic process | (none in phylogroup) | 1 |
| 0 - ALL | Process | organelle assembly | 0.335363566 | 1 |
| 0 - ALL | Process | organelle fission | -1.629012524 | 0.8612394 |
| 0 - ALL | Process | organelle fusion | -1.673406644 | 1 |
| 0 - ALL | Process | organelle inheritance | 0.670547757 | 1 |
| 0 - ALL | Process | peptidyl-amino acid modification | -0.291977537 | 1 |
| 0 - ALL | Process | peroxisome organization | -1.716475366 | 1 |
| 0 - ALL | Process | protein acylation | -0.021329947 | 1 |
| 0 - ALL | Process | protein alkylation | (none in phylogroup) | 1 |
| 0 - ALL | Process | protein complex biogenesis | -2.363722145 | 0.05161777 |
| 0 - ALL | Process | protein dephosphorylation | -0.876940038 | 1 |
| 0 - ALL | Process | protein folding | 0.496518358 | 1 |
| 0 - ALL | Process | protein glycosylation | 0.262463019 | 1 |
| 0 - ALL | Process | protein lipidation | -0.055277279 | 1 |
| 0 - ALL | Process | protein maturation | 0.085585257 | 1 |
| 0 - ALL | Process | protein modification by small protein conjugation or removal | -2.808921614 | 0.1571312 |
| 0 - ALL | Process | protein phosphorylation | (none in phylogroup) | 0.01894893 |
| 0 - ALL | Process | protein targeting | -0.844025415 | 0.9764787 |
| 0 - ALL | Process | proteolysis involved in cellular protein catabolic process | -3.198868133 | 0.02713984 |
| 0 - ALL | Process | pseudohyphal growth | -0.110139214 | 1 |
| 0 - ALL | Process | regulation of cell cycle | -3.175906984 | 0.03067461 |
| 0 - ALL | Process | regulation of DNA metabolic process | 0.416791165 | 1 |
| 0 - ALL | Process | regulation of organelle organization | -0.662435526 | 1 |
| 0 - ALL | Process | regulation of protein modification process | (none in phylogroup) | 0.7928933 |
| 0 - ALL | Process | regulation of translation | -0.48699352 | 1 |
| 0 - ALL | Process | regulation of transport | (none in phylogroup) | 0.971396 |
| 0 - ALL | Process | response to chemical stimulus | 0.213773526 | 1 |
| 0 - ALL | Process | response to DNA damage stimulus | -0.446635782 | 1 |
| 0 - ALL | Process | response to heat | -0.914414743 | 1 |
| 0 - ALL | Process | response to osmotic stress | -0.233874583 | 1 |
| 0 - ALL | Process | response to oxidative stress | 2.049059381 | 6.79E-06 |
| 0 - ALL | Process | response to starvation | 0.142168785 | 1 |
| 0 - ALL | Process | ribosomal large subunit biogenesis | 0.013435471 | 1 |
| 0 - ALL | Process | ribosomal small subunit biogenesis | -0.26128074 | 1 |
| 0 - ALL | Process | ribosomal subunit export from nucleus | (none in phylogroup) | 1 |
| 0 - ALL | Process | ribosome assembly | 0.810272521 | 1 |
| 0 - ALL | Process | RNA catabolic process | -0.370479511 | 1 |
| 0 - ALL | Process | RNA modification | 0.911555857 | 0.1847382 |
| 0 - ALL | Process | RNA splicing | -1.727044607 | 0.6716412 |
| 0 - ALL | Process | rRNA processing | -0.359405465 | 1 |
| 0 - ALL | Process | signaling | -2.474502575 | 0.02728584 |
| 0 - ALL | Process | snoRNA processing | 0.868487135 | 1 |
| 0 - ALL | Process | sporulation | -1.617697211 | 0.8836356 |
| 0 - ALL | Process | telomere organization | -0.088444143 | 1 |
| 0 - ALL | Process | transcription elongation, DNA-dependent | (none in phylogroup) | 0.6094631 |
| 0 - ALL | Process | transcription from RNA polymerase I promoter | (none in phylogroup) | 0.971396 |
| 0 - ALL | Process | transcription from RNA polymerase II promoter | -2.356932979 | 0.000292915 |
| 0 - ALL | Process | transcription from RNA polymerase III promoter | -0.950940619 | 1 |
| 0 - ALL | Process | transcription initiation, DNA-dependent | (none in phylogroup) | 0.5702022 |
| 0 - ALL | Process | transcription termination, DNA-dependent | 0.46409688 | 1 |
| 0 - ALL | Process | translational elongation | -2.999699931 | 0.000474207 |
| 0 - ALL | Process | translational initiation | (none in phylogroup) | 1 |
| 0 - ALL | Process | transmembrane transport | -0.183601376 | 1 |
| 0 - ALL | Process | transposition | (none in phylogroup) | 0.1265165 |
| 0 - ALL | Process | tRNA aminoacylation for protein translation | 3.331459111 | 0 |
| 0 - ALL | Process | tRNA processing | 1.21570925 | 0.07524597 |
| 0 - ALL | Process | vacuole organization | -0.329452243 | 1 |
| 0 - ALL | Process | vesicle organization | -1.895799065 | 1 |
| 0 - ALL | Process | vitamin metabolic process | 1.138326719 | 0.5135718 |
| 1 - ALL (-ARCHAEA) | Component | cell cortex | (none in phylogroup) | 0.0559386 |
| 1 - ALL (-ARCHAEA) | Component | cell wall | -0.530848786 | 1 |
| 1 - ALL (-ARCHAEA) | Component | cellular bud | (none in phylogroup) | 0.008630738 |
| 1 - ALL (-ARCHAEA) | Component | cellular component unknown | -0.751786708 | 0.1507798 |
| 1 - ALL (-ARCHAEA) | Component | chromosome | -2.344563873 | 0.004721203 |
| 1 - ALL (-ARCHAEA) | Component | cytoplasm | 0.500658488 | 0 |
| 1 - ALL (-ARCHAEA) | Component | cytoplasmic membrane-bounded vesicle | (none in phylogroup) | 0.1841856 |
| 1 - ALL (-ARCHAEA) | Component | cytoskeleton | (none in phylogroup) | 0.002733167 |
| 1 - ALL (-ARCHAEA) | Component | endomembrane system | -1.188323786 | 0.1953234 |
| 1 - ALL (-ARCHAEA) | Component | endoplasmic reticulum | -0.760024388 | 0.774286 |
| 1 - ALL (-ARCHAEA) | Component | extracellular region | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Component | Golgi apparatus | -1.470094755 | 0.5983276 |
| 1 - ALL (-ARCHAEA) | Component | membrane | -0.051550094 | 1 |
| 1 - ALL (-ARCHAEA) | Component | membrane fraction | 0.790614598 | 0.2605722 |
| 1 - ALL (-ARCHAEA) | Component | microtubule organizing center | (none in phylogroup) | 0.6728817 |
| 1 - ALL (-ARCHAEA) | Component | mitochondrial envelope | 1.583936823 | 1.41E-09 |
| 1 - ALL (-ARCHAEA) | Component | mitochondrion | 1.544222548 | 0 |
| 1 - ALL (-ARCHAEA) | Component | nucleolus | -1.243818899 | 0.2385761 |
| 1 - ALL (-ARCHAEA) | Component | nucleus | -0.758293084 | 1.62E-05 |
| 1 - ALL (-ARCHAEA) | Component | peroxisome | 1.461733743 | 0.06766475 |
| 1 - ALL (-ARCHAEA) | Component | plasma membrane | -1.368896032 | 0.2212349 |
| 1 - ALL (-ARCHAEA) | Component | ribosome | 0.931560533 | 0.008159774 |
| 1 - ALL (-ARCHAEA) | Component | site of polarized growth | -3.342446932 | 0.01222545 |
| 1 - ALL (-ARCHAEA) | Component | vacuole | -0.994870249 | 0.999019 |
| 1 - ALL (-ARCHAEA) | Function | ATPase activity | 1.9605396 | 1.32E-12 |
| 1 - ALL (-ARCHAEA) | Function | cytoskeletal protein binding | (none in phylogroup) | 0.8836356 |
| 1 - ALL (-ARCHAEA) | Function | DNA binding | -0.611822864 | 1 |
| 1 - ALL (-ARCHAEA) | Function | enzyme binding | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Function | enzyme regulator activity | -3.284082769 | 0.01686778 |
| 1 - ALL (-ARCHAEA) | Function | GTPase activity | 1.236174042 | 0.2998789 |
| 1 - ALL (-ARCHAEA) | Function | helicase activity | 1.162173461 | 0.2218101 |
| 1 - ALL (-ARCHAEA) | Function | histone binding | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Function | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 0.875292313 | 0.9232297 |
| 1 - ALL (-ARCHAEA) | Function | hydrolase activity, acting on glycosyl bonds | 2.099437705 | 0.001616365 |
| 1 - ALL (-ARCHAEA) | Function | ion binding | 0.711512052 | 1 |
| 1 - ALL (-ARCHAEA) | Function | isomerase activity | 2.11064316 | 0.000342435 |
| 1 - ALL (-ARCHAEA) | Function | kinase activity | 0.285321692 | 1 |
| 1 - ALL (-ARCHAEA) | Function | ligase activity | 0.692688177 | 0.602739 |
| 1 - ALL (-ARCHAEA) | Function | lipid binding | (none in phylogroup) | 0.4027664 |
| 1 - ALL (-ARCHAEA) | Function | lyase activity | 0.109062124 | 1 |
| 1 - ALL (-ARCHAEA) | Function | methyltransferase activity | 0.865191723 | 0.7040205 |
| 1 - ALL (-ARCHAEA) | Function | molecular function unknown | -1.526043002 | 1.71E-14 |
| 1 - ALL (-ARCHAEA) | Function | mRNA binding | 0.236174042 | 1 |
| 1 - ALL (-ARCHAEA) | Function | nuclease activity | 0.046696243 | 1 |
| 1 - ALL (-ARCHAEA) | Function | nucleic acid binding transcription factor activity | (none in phylogroup) | 0.4354082 |
| 1 - ALL (-ARCHAEA) | Function | nucleotidyltransferase activity | -1.336077407 | 1 |
| 1 - ALL (-ARCHAEA) | Function | oxidoreductase activity | 1.9605396 | 0 |
| 1 - ALL (-ARCHAEA) | Function | peptidase activity | 0.707997568 | 0.774286 |
| 1 - ALL (-ARCHAEA) | Function | phosphatase activity | -0.530848786 | 1 |
| 1 - ALL (-ARCHAEA) | Function | protein binding transcription factor activity | -1.763825958 | 1 |
| 1 - ALL (-ARCHAEA) | Function | protein binding, bridging | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Function | protein transporter activity | -1.188323786 | 1 |
| 1 - ALL (-ARCHAEA) | Function | RNA binding | -0.904394156 | 0.03803129 |
| 1 - ALL (-ARCHAEA) | Function | RNA modification guide activity | (none in phylogroup) | 0.7154833 |
| 1 - ALL (-ARCHAEA) | Function | rRNA binding | (none in phylogroup) | 0.3157589 |
| 1 - ALL (-ARCHAEA) | Function | signal transducer activity | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Function | small conjugating protein binding | -0.942163199 | 1 |
| 1 - ALL (-ARCHAEA) | Function | structural constituent of ribosome | 1.005129751 | 0.03131794 |
| 1 - ALL (-ARCHAEA) | Function | structural molecule activity | 0.3384241 | 1 |
| 1 - ALL (-ARCHAEA) | Function | transcription factor binding | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Function | transferase activity, transferring acyl groups | 1.261709134 | 0.04510092 |
| 1 - ALL (-ARCHAEA) | Function | transferase activity, transferring alkyl or aryl (other than methyl) groups | 1.520627432 | 0.1819127 |
| 1 - ALL (-ARCHAEA) | Function | transferase activity, transferring glycosyl groups | -1.159754634 | 1 |
| 1 - ALL (-ARCHAEA) | Function | translation factor activity, nucleic acid binding | 2.130464601 | 0.001356743 |
| 1 - ALL (-ARCHAEA) | Function | transmembrane transporter activity | 0.676746634 | 0.2469689 |
| 1 - ALL (-ARCHAEA) | Function | triplet codon-amino acid adaptor activity | (none in phylogroup) | 5.86E-05 |
| 1 - ALL (-ARCHAEA) | Function | unfolded protein binding | 1.002974866 | 0.586864 |
| 1 - ALL (-ARCHAEA) | Process | amino acid transport | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Process | biological process unknown | -1.006563648 | 0.000263718 |
| 1 - ALL (-ARCHAEA) | Process | carbohydrate metabolic process | 1.595522838 | 2.57E-08 |
| 1 - ALL (-ARCHAEA) | Process | carbohydrate transport | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Process | cell budding | (none in phylogroup) | 0.4354082 |
| 1 - ALL (-ARCHAEA) | Process | cell morphogenesis | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Process | cell wall organization or biogenesis | -1.705723003 | 0.2405581 |
| 1 - ALL (-ARCHAEA) | Process | cellular amino acid metabolic process | 1.277650678 | 0.000409947 |
| 1 - ALL (-ARCHAEA) | Process | cellular ion homeostasis | 0.711512052 | 0.850762 |
| 1 - ALL (-ARCHAEA) | Process | cellular respiration | 0.9605396 | 0.455073 |
| 1 - ALL (-ARCHAEA) | Process | chromatin organization | -0.962154674 | 1 |
| 1 - ALL (-ARCHAEA) | Process | chromosome segregation | -1.592714041 | 0.9347144 |
| 1 - ALL (-ARCHAEA) | Process | cofactor metabolic process | 2.429653772 | 0 |
| 1 - ALL (-ARCHAEA) | Process | conjugation | (none in phylogroup) | 0.1265165 |
| 1 - ALL (-ARCHAEA) | Process | cytokinesis | -1.297258158 | 1 |
| 1 - ALL (-ARCHAEA) | Process | cytoplasmic translation | -0.95884194 | 1 |
| 1 - ALL (-ARCHAEA) | Process | cytoskeleton organization | -3.303801004 | 0.0150926 |
| 1 - ALL (-ARCHAEA) | Process | DNA recombination | -1.130608288 | 1 |
| 1 - ALL (-ARCHAEA) | Process | DNA repair | -0.554033573 | 1 |
| 1 - ALL (-ARCHAEA) | Process | DNA replication | -0.302494806 | 1 |
| 1 - ALL (-ARCHAEA) | Process | endocytosis | (none in phylogroup) | 0.2833653 |
| 1 - ALL (-ARCHAEA) | Process | endosome transport | (none in phylogroup) | 0.6260293 |
| 1 - ALL (-ARCHAEA) | Process | exocytosis | -1.070487296 | 1 |
| 1 - ALL (-ARCHAEA) | Process | generation of precursor metabolites and energy | 1.365706855 | 0.001975812 |
| 1 - ALL (-ARCHAEA) | Process | Golgi vesicle transport | (none in phylogroup) | 0.008630738 |
| 1 - ALL (-ARCHAEA) | Process | histone modification | (none in phylogroup) | 0.2833653 |
| 1 - ALL (-ARCHAEA) | Process | invasive growth in response to glucose limitation | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Process | ion transport | 1.450678554 | 0.003042996 |
| 1 - ALL (-ARCHAEA) | Process | lipid metabolic process | 0.258033476 | 1 |
| 1 - ALL (-ARCHAEA) | Process | lipid transport | -0.130608288 | 1 |
| 1 - ALL (-ARCHAEA) | Process | meiotic cell cycle | -0.864626599 | 1 |
| 1 - ALL (-ARCHAEA) | Process | membrane fusion | (none in phylogroup) | 0.5702022 |
| 1 - ALL (-ARCHAEA) | Process | membrane invagination | (none in phylogroup) | 0.05826233 |
| 1 - ALL (-ARCHAEA) | Process | mitochondrial translation | 2.314176554 | 2.59E-12 |
| 1 - ALL (-ARCHAEA) | Process | mitochondrion organization | 1.631658744 | 1.54E-10 |
| 1 - ALL (-ARCHAEA) | Process | mitotic cell cycle | -1.665645564 | 0.1004714 |
| 1 - ALL (-ARCHAEA) | Process | mRNA processing | -0.677786127 | 1 |
| 1 - ALL (-ARCHAEA) | Process | nuclear transport | -1.234716692 | 1 |
| 1 - ALL (-ARCHAEA) | Process | nucleobase, nucleoside and nucleotide metabolic process | 2.004933719 | 9.89E-11 |
| 1 - ALL (-ARCHAEA) | Process | nucleobase, nucleoside, nucleotide and nucleic acid transport | -1.348788459 | 1 |
| 1 - ALL (-ARCHAEA) | Process | nucleus organization | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Process | oligosaccharide metabolic process | 2.596576285 | 2.72E-05 |
| 1 - ALL (-ARCHAEA) | Process | organelle assembly | (none in phylogroup) | 0.4880645 |
| 1 - ALL (-ARCHAEA) | Process | organelle fission | -1.515898444 | 1 |
| 1 - ALL (-ARCHAEA) | Process | organelle fusion | (none in phylogroup) | 0.8567433 |
| 1 - ALL (-ARCHAEA) | Process | organelle inheritance | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Process | peptidyl-amino acid modification | (none in phylogroup) | 0.607116 |
| 1 - ALL (-ARCHAEA) | Process | peroxisome organization | -1.603361286 | 1 |
| 1 - ALL (-ARCHAEA) | Process | protein acylation | (none in phylogroup) | 0.9389088 |
| 1 - ALL (-ARCHAEA) | Process | protein alkylation | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Process | protein complex biogenesis | -0.250608065 | 1 |
| 1 - ALL (-ARCHAEA) | Process | protein dephosphorylation | -0.763825958 | 1 |
| 1 - ALL (-ARCHAEA) | Process | protein folding | 1.194594938 | 0.1501669 |
| 1 - ALL (-ARCHAEA) | Process | protein glycosylation | -1.624422901 | 1 |
| 1 - ALL (-ARCHAEA) | Process | protein lipidation | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Process | protein maturation | 2.006054259 | 0.01070551 |
| 1 - ALL (-ARCHAEA) | Process | protein modification by small protein conjugation or removal | -2.695807534 | 0.2385761 |
| 1 - ALL (-ARCHAEA) | Process | protein phosphorylation | (none in phylogroup) | 0.03153485 |
| 1 - ALL (-ARCHAEA) | Process | protein targeting | -0.368341256 | 1 |
| 1 - ALL (-ARCHAEA) | Process | proteolysis involved in cellular protein catabolic process | -0.278399131 | 1 |
| 1 - ALL (-ARCHAEA) | Process | pseudohyphal growth | (none in phylogroup) | 0.829418 |
| 1 - ALL (-ARCHAEA) | Process | regulation of cell cycle | -2.062792904 | 0.2143507 |
| 1 - ALL (-ARCHAEA) | Process | regulation of DNA metabolic process | 0.529905245 | 1 |
| 1 - ALL (-ARCHAEA) | Process | regulation of organelle organization | -1.549321446 | 1 |
| 1 - ALL (-ARCHAEA) | Process | regulation of protein modification process | (none in phylogroup) | 0.9389088 |
| 1 - ALL (-ARCHAEA) | Process | regulation of translation | 0.363086155 | 1 |
| 1 - ALL (-ARCHAEA) | Process | regulation of transport | -1.348788459 | 1 |
| 1 - ALL (-ARCHAEA) | Process | response to chemical stimulus | 0.602522049 | 0.3640214 |
| 1 - ALL (-ARCHAEA) | Process | response to DNA damage stimulus | -0.655449797 | 1 |
| 1 - ALL (-ARCHAEA) | Process | response to heat | 0.198699337 | 1 |
| 1 - ALL (-ARCHAEA) | Process | response to osmotic stress | -0.120760503 | 1 |
| 1 - ALL (-ARCHAEA) | Process | response to oxidative stress | 2.07471062 | 1.20E-05 |
| 1 - ALL (-ARCHAEA) | Process | response to starvation | (none in phylogroup) | 0.6260293 |
| 1 - ALL (-ARCHAEA) | Process | ribosomal large subunit biogenesis | -0.873450449 | 1 |
| 1 - ALL (-ARCHAEA) | Process | ribosomal small subunit biogenesis | -1.470094755 | 1 |
| 1 - ALL (-ARCHAEA) | Process | ribosomal subunit export from nucleus | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Process | ribosome assembly | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Process | RNA catabolic process | 0.064562664 | 1 |
| 1 - ALL (-ARCHAEA) | Process | RNA modification | 0.676746634 | 0.774286 |
| 1 - ALL (-ARCHAEA) | Process | RNA splicing | -0.292002432 | 1 |
| 1 - ALL (-ARCHAEA) | Process | rRNA processing | -0.705723003 | 1 |
| 1 - ALL (-ARCHAEA) | Process | signaling | -1.776425995 | 0.17328 |
| 1 - ALL (-ARCHAEA) | Process | snoRNA processing | 0.396638714 | 1 |
| 1 - ALL (-ARCHAEA) | Process | sporulation | (none in phylogroup) | 0.08342663 |
| 1 - ALL (-ARCHAEA) | Process | telomere organization | 0.024669937 | 1 |
| 1 - ALL (-ARCHAEA) | Process | transcription elongation, DNA-dependent | -1.645181461 | 1 |
| 1 - ALL (-ARCHAEA) | Process | transcription from RNA polymerase I promoter | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Process | transcription from RNA polymerase II promoter | -2.243818899 | 0.001034915 |
| 1 - ALL (-ARCHAEA) | Process | transcription from RNA polymerase III promoter | (none in phylogroup) | 1 |
| 1 - ALL (-ARCHAEA) | Process | transcription initiation, DNA-dependent | (none in phylogroup) | 0.6918753 |
| 1 - ALL (-ARCHAEA) | Process | transcription termination, DNA-dependent | -0.42278904 | 1 |
| 1 - ALL (-ARCHAEA) | Process | translational elongation | -1.564657756 | 0.06924424 |
| 1 - ALL (-ARCHAEA) | Process | translational initiation | 1.484101556 | 0.09228123 |
| 1 - ALL (-ARCHAEA) | Process | transmembrane transport | 1.016975545 | 0.05511774 |
| 1 - ALL (-ARCHAEA) | Process | transposition | (none in phylogroup) | 0.1771223 |
| 1 - ALL (-ARCHAEA) | Process | tRNA aminoacylation for protein translation | 2.27464819 | 0.001331447 |
| 1 - ALL (-ARCHAEA) | Process | tRNA processing | 1.32882333 | 0.04307133 |
| 1 - ALL (-ARCHAEA) | Process | vacuole organization | (none in phylogroup) | 0.07356634 |
| 1 - ALL (-ARCHAEA) | Process | vesicle organization | (none in phylogroup) | 0.5864367 |
| 1 - ALL (-ARCHAEA) | Process | vitamin metabolic process | 1.514475205 | 0.1253657 |
| 2 - ALL (-BACTERIA) | Component | cell cortex | -2.393242412 | 0.5892075 |
| 2 - ALL (-BACTERIA) | Component | cell wall | (none in phylogroup) | 0.4694521 |
| 2 - ALL (-BACTERIA) | Component | cellular bud | (none in phylogroup) | 0.02485965 |
| 2 - ALL (-BACTERIA) | Component | cellular component unknown | -3.779026106 | 8.04E-09 |
| 2 - ALL (-BACTERIA) | Component | chromosome | 0.098516664 | 1 |
| 2 - ALL (-BACTERIA) | Component | cytoplasm | 0.423167111 | 1.13E-10 |
| 2 - ALL (-BACTERIA) | Component | cytoplasmic membrane-bounded vesicle | (none in phylogroup) | 0.3326457 |
| 2 - ALL (-BACTERIA) | Component | cytoskeleton | -3.009455847 | 0.06924424 |
| 2 - ALL (-BACTERIA) | Component | endomembrane system | 0.131900002 | 1 |
| 2 - ALL (-BACTERIA) | Component | endoplasmic reticulum | -0.053909445 | 1 |
| 2 - ALL (-BACTERIA) | Component | extracellular region | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Component | Golgi apparatus | (none in phylogroup) | 0.0223337 |
| 2 - ALL (-BACTERIA) | Component | membrane | -1.261496334 | 3.31E-06 |
| 2 - ALL (-BACTERIA) | Component | membrane fraction | -2.988697287 | 0.07538048 |
| 2 - ALL (-BACTERIA) | Component | microtubule organizing center | (none in phylogroup) | 0.95354 |
| 2 - ALL (-BACTERIA) | Component | mitochondrial envelope | -2.621639816 | 0.01074926 |
| 2 - ALL (-BACTERIA) | Component | mitochondrion | -1.434761682 | 1.60E-05 |
| 2 - ALL (-BACTERIA) | Component | nucleolus | 0.915468672 | 0.03568229 |
| 2 - ALL (-BACTERIA) | Component | nucleus | 0.140466936 | 1 |
| 2 - ALL (-BACTERIA) | Component | peroxisome | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Component | plasma membrane | -3.470136012 | 0.005533816 |
| 2 - ALL (-BACTERIA) | Component | ribosome | 3.052712974 | 0 |
| 2 - ALL (-BACTERIA) | Component | site of polarized growth | -3.121758817 | 0.04111517 |
| 2 - ALL (-BACTERIA) | Component | vacuole | -0.511147728 | 1 |
| 2 - ALL (-BACTERIA) | Function | ATPase activity | 1.181227715 | 0.00762123 |
| 2 - ALL (-BACTERIA) | Function | cytoskeletal protein binding | -0.317578142 | 1 |
| 2 - ALL (-BACTERIA) | Function | DNA binding | -0.012623126 | 1 |
| 2 - ALL (-BACTERIA) | Function | enzyme binding | -0.580612548 | 1 |
| 2 - ALL (-BACTERIA) | Function | enzyme regulator activity | -0.741466559 | 1 |
| 2 - ALL (-BACTERIA) | Function | GTPase activity | 0.456862157 | 1 |
| 2 - ALL (-BACTERIA) | Function | helicase activity | 1.382861576 | 0.09649024 |
| 2 - ALL (-BACTERIA) | Function | histone binding | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Function | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 0.359014834 | 1 |
| 2 - ALL (-BACTERIA) | Function | hydrolase activity, acting on glycosyl bonds | 0.150200819 | 1 |
| 2 - ALL (-BACTERIA) | Function | ion binding | -0.652762334 | 1 |
| 2 - ALL (-BACTERIA) | Function | isomerase activity | -1.128100343 | 1 |
| 2 - ALL (-BACTERIA) | Function | kinase activity | -0.631493717 | 1 |
| 2 - ALL (-BACTERIA) | Function | ligase activity | 0.020291497 | 1 |
| 2 - ALL (-BACTERIA) | Function | lipid binding | -1.738153825 | 1 |
| 2 - ALL (-BACTERIA) | Function | lyase activity | -0.08528726 | 1 |
| 2 - ALL (-BACTERIA) | Function | methyltransferase activity | 1.448449918 | 0.05009132 |
| 2 - ALL (-BACTERIA) | Function | molecular function unknown | -1.674588696 | 7.49E-14 |
| 2 - ALL (-BACTERIA) | Function | mRNA binding | -1.543137843 | 1 |
| 2 - ALL (-BACTERIA) | Function | nuclease activity | 0.004349953 | 1 |
| 2 - ALL (-BACTERIA) | Function | nucleic acid binding transcription factor activity | (none in phylogroup) | 0.6746366 |
| 2 - ALL (-BACTERIA) | Function | nucleotidyltransferase activity | 1.469573209 | 0.01380974 |
| 2 - ALL (-BACTERIA) | Function | oxidoreductase activity | -1.818772285 | 0.142435 |
| 2 - ALL (-BACTERIA) | Function | peptidase activity | -0.808279911 | 1 |
| 2 - ALL (-BACTERIA) | Function | phosphatase activity | (none in phylogroup) | 0.4694521 |
| 2 - ALL (-BACTERIA) | Function | protein binding transcription factor activity | 0.041824658 | 1 |
| 2 - ALL (-BACTERIA) | Function | protein binding, bridging | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Function | protein transporter activity | 0.032364329 | 1 |
| 2 - ALL (-BACTERIA) | Function | RNA binding | 0.468297052 | 0.2858688 |
| 2 - ALL (-BACTERIA) | Function | RNA modification guide activity | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Function | rRNA binding | -0.24940664 | 1 |
| 2 - ALL (-BACTERIA) | Function | signal transducer activity | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Function | small conjugating protein binding | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Function | structural constituent of ribosome | 3.443048582 | 0 |
| 2 - ALL (-BACTERIA) | Function | structural molecule activity | 2.776342932 | 0 |
| 2 - ALL (-BACTERIA) | Function | transcription factor binding | -0.543137843 | 1 |
| 2 - ALL (-BACTERIA) | Function | transferase activity, transferring acyl groups | -1.102565251 | 1 |
| 2 - ALL (-BACTERIA) | Function | transferase activity, transferring alkyl or aryl (other than methyl) groups | 0.419387452 | 1 |
| 2 - ALL (-BACTERIA) | Function | transferase activity, transferring glycosyl groups | -0.354104018 | 1 |
| 2 - ALL (-BACTERIA) | Function | translation factor activity, nucleic acid binding | 2.988582637 | 8.01E-09 |
| 2 - ALL (-BACTERIA) | Function | transmembrane transporter activity | -0.977034369 | 0.7994005 |
| 2 - ALL (-BACTERIA) | Function | triplet codon-amino acid adaptor activity | (none in phylogroup) | 0.000363075 |
| 2 - ALL (-BACTERIA) | Function | unfolded protein binding | 0.223662981 | 1 |
| 2 - ALL (-BACTERIA) | Process | amino acid transport | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | biological process unknown | -2.370838034 | 5.47E-11 |
| 2 - ALL (-BACTERIA) | Process | carbohydrate metabolic process | -0.808279911 | 1 |
| 2 - ALL (-BACTERIA) | Process | carbohydrate transport | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | cell budding | (none in phylogroup) | 0.6746366 |
| 2 - ALL (-BACTERIA) | Process | cell morphogenesis | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | cell wall organization or biogenesis | (none in phylogroup) | 0.007205446 |
| 2 - ALL (-BACTERIA) | Process | cellular amino acid metabolic process | 0.11982717 | 1 |
| 2 - ALL (-BACTERIA) | Process | cellular ion homeostasis | 0.08420326 | 1 |
| 2 - ALL (-BACTERIA) | Process | cellular respiration | -1.818772285 | 1 |
| 2 - ALL (-BACTERIA) | Process | chromatin organization | -1.478432153 | 0.5864367 |
| 2 - ALL (-BACTERIA) | Process | chromosome segregation | -0.787063426 | 1 |
| 2 - ALL (-BACTERIA) | Process | cofactor metabolic process | -1.050097831 | 1 |
| 2 - ALL (-BACTERIA) | Process | conjugation | -2.165575049 | 0.9550368 |
| 2 - ALL (-BACTERIA) | Process | cytokinesis | -1.076570043 | 1 |
| 2 - ALL (-BACTERIA) | Process | cytoplasmic translation | 3.816435027 | 0 |
| 2 - ALL (-BACTERIA) | Process | cytoskeleton organization | -3.083112889 | 0.04985459 |
| 2 - ALL (-BACTERIA) | Process | DNA recombination | 1.090079827 | 0.1155876 |
| 2 - ALL (-BACTERIA) | Process | DNA repair | 0.444262121 | 1 |
| 2 - ALL (-BACTERIA) | Process | DNA replication | 1.296704932 | 0.02713984 |
| 2 - ALL (-BACTERIA) | Process | endocytosis | -1.88017283 | 1 |
| 2 - ALL (-BACTERIA) | Process | endosome transport | -1.52402902 | 1 |
| 2 - ALL (-BACTERIA) | Process | exocytosis | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | generation of precursor metabolites and energy | -2.661532543 | 0.2675034 |
| 2 - ALL (-BACTERIA) | Process | Golgi vesicle transport | -2.810910168 | 0.1571599 |
| 2 - ALL (-BACTERIA) | Process | histone modification | -0.88017283 | 1 |
| 2 - ALL (-BACTERIA) | Process | invasive growth in response to glucose limitation | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | ion transport | -2.328633331 | 0.6820937 |
| 2 - ALL (-BACTERIA) | Process | lipid metabolic process | -0.521278409 | 1 |
| 2 - ALL (-BACTERIA) | Process | lipid transport | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | meiotic cell cycle | -0.058975983 | 1 |
| 2 - ALL (-BACTERIA) | Process | membrane fusion | -1.580612548 | 1 |
| 2 - ALL (-BACTERIA) | Process | membrane invagination | -1.38267317 | 1 |
| 2 - ALL (-BACTERIA) | Process | mitochondrial translation | (none in phylogroup) | 0.132819 |
| 2 - ALL (-BACTERIA) | Process | mitochondrion organization | (none in phylogroup) | 0.000136695 |
| 2 - ALL (-BACTERIA) | Process | mitotic cell cycle | -0.123029354 | 1 |
| 2 - ALL (-BACTERIA) | Process | mRNA processing | 0.028328816 | 1 |
| 2 - ALL (-BACTERIA) | Process | nuclear transport | 1.488471764 | 0.00151479 |
| 2 - ALL (-BACTERIA) | Process | nucleobase, nucleoside and nucleotide metabolic process | -0.49684419 | 1 |
| 2 - ALL (-BACTERIA) | Process | nucleobase, nucleoside, nucleotide and nucleic acid transport | 1.456862157 | 0.01473155 |
| 2 - ALL (-BACTERIA) | Process | nucleus organization | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | oligosaccharide metabolic process | 0.495336305 | 1 |
| 2 - ALL (-BACTERIA) | Process | organelle assembly | 2.254128262 | 5.14E-06 |
| 2 - ALL (-BACTERIA) | Process | organelle fission | -1.295210329 | 1 |
| 2 - ALL (-BACTERIA) | Process | organelle fusion | -1.339604449 | 1 |
| 2 - ALL (-BACTERIA) | Process | organelle inheritance | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | peptidyl-amino acid modification | 0.041824658 | 1 |
| 2 - ALL (-BACTERIA) | Process | peroxisome organization | -1.38267317 | 1 |
| 2 - ALL (-BACTERIA) | Process | protein acylation | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | protein alkylation | -0.617138424 | 1 |
| 2 - ALL (-BACTERIA) | Process | protein complex biogenesis | -0.444957449 | 1 |
| 2 - ALL (-BACTERIA) | Process | protein dephosphorylation | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | protein folding | -0.169679447 | 1 |
| 2 - ALL (-BACTERIA) | Process | protein glycosylation | 0.596265214 | 1 |
| 2 - ALL (-BACTERIA) | Process | protein lipidation | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | protein maturation | 0.419387452 | 1 |
| 2 - ALL (-BACTERIA) | Process | protein modification by small protein conjugation or removal | -2.475119419 | 0.4782647 |
| 2 - ALL (-BACTERIA) | Process | protein phosphorylation | -0.533615069 | 1 |
| 2 - ALL (-BACTERIA) | Process | protein targeting | -0.995650047 | 0.998156 |
| 2 - ALL (-BACTERIA) | Process | proteolysis involved in cellular protein catabolic process | -0.057711015 | 1 |
| 2 - ALL (-BACTERIA) | Process | pseudohyphal growth | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | regulation of cell cycle | -1.257142288 | 1 |
| 2 - ALL (-BACTERIA) | Process | regulation of DNA metabolic process | 1.072521455 | 0.6094631 |
| 2 - ALL (-BACTERIA) | Process | regulation of organelle organization | (none in phylogroup) | 0.1507798 |
| 2 - ALL (-BACTERIA) | Process | regulation of protein modification process | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | regulation of translation | 2.261846175 | 2.06E-06 |
| 2 - ALL (-BACTERIA) | Process | regulation of transport | (none in phylogroup) | 1 |
| 2 - ALL (-BACTERIA) | Process | response to chemical stimulus | -2.115389292 | 0.02530315 |
| 2 - ALL (-BACTERIA) | Process | response to DNA damage stimulus | 0.150200819 | 1 |
| 2 - ALL (-BACTERIA) | Process | response to heat | -0.580612548 | 1 |
| 2 - ALL (-BACTERIA) | Process | response to osmotic stress | (none in phylogroup) | 0.95354 |
| 2 - ALL (-BACTERIA) | Process | response to oxidative stress | -1.704601265 | 1 |
| 2 - ALL (-BACTERIA) | Process | response to starvation | 0.060933481 | 1 |
| 2 - ALL (-BACTERIA) | Process | ribosomal large subunit biogenesis | 2.047677384 | 0.000138447 |
| 2 - ALL (-BACTERIA) | Process | ribosomal small subunit biogenesis | 2.960046726 | 0 |
| 2 - ALL (-BACTERIA) | Process | ribosomal subunit export from nucleus | 1.634400343 | 0.1340566 |
| 2 - ALL (-BACTERIA) | Process | ribosome assembly | 2.729037217 | 3.71E-08 |
| 2 - ALL (-BACTERIA) | Process | RNA catabolic process | 0.963322684 | 0.455073 |
| 2 - ALL (-BACTERIA) | Process | RNA modification | 1.138442848 | 0.068228 |
| 2 - ALL (-BACTERIA) | Process | RNA splicing | 0.41411251 | 1 |
| 2 - ALL (-BACTERIA) | Process | rRNA processing | 2.038527068 | 0 |
| 2 - ALL (-BACTERIA) | Process | signaling | -2.14070038 | 0.1542483 |
| 2 - ALL (-BACTERIA) | Process | snoRNA processing | 1.61732683 | 0.2213474 |
| 2 - ALL (-BACTERIA) | Process | sporulation | (none in phylogroup) | 0.1715982 |
| 2 - ALL (-BACTERIA) | Process | telomere organization | -0.339604449 | 1 |
| 2 - ALL (-BACTERIA) | Process | transcription elongation, DNA-dependent | -1.424493346 | 1 |
| 2 - ALL (-BACTERIA) | Process | transcription from RNA polymerase I promoter | 1.456862157 | 0.1527761 |
| 2 - ALL (-BACTERIA) | Process | transcription from RNA polymerase II promoter | 0.064332057 | 1 |
| 2 - ALL (-BACTERIA) | Process | transcription from RNA polymerase III promoter | 2.382861576 | 0.000810666 |
| 2 - ALL (-BACTERIA) | Process | transcription initiation, DNA-dependent | 0.856792764 | 0.9562792 |
| 2 - ALL (-BACTERIA) | Process | transcription termination, DNA-dependent | 1.382861576 | 0.5484764 |
| 2 - ALL (-BACTERIA) | Process | translational elongation | 0.504027265 | 0.8809765 |
| 2 - ALL (-BACTERIA) | Process | translational initiation | 1.897434749 | 0.009879267 |
| 2 - ALL (-BACTERIA) | Process | transmembrane transport | -2.849799181 | 0.1361538 |
| 2 - ALL (-BACTERIA) | Process | transposition | -0.465135331 | 1 |
| 2 - ALL (-BACTERIA) | Process | tRNA aminoacylation for protein translation | 2.302691227 | 0.002889855 |
| 2 - ALL (-BACTERIA) | Process | tRNA processing | 1.675042327 | 0.00382814 |
| 2 - ALL (-BACTERIA) | Process | vacuole organization | -2.317578142 | 0.6975441 |
| 2 - ALL (-BACTERIA) | Process | vesicle organization | -1.56199687 | 1 |
| 2 - ALL (-BACTERIA) | Process | vitamin metabolic process | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Component | cell cortex | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Component | cell wall | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Component | cellular bud | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Component | cellular component unknown | 0.05237429 | 1 |
| 3 - ALL (-ANIMALS) | Component | chromosome | (none in phylogroup) | 0.971396 |
| 3 - ALL (-ANIMALS) | Component | cytoplasm | 0.224820164 | 0.8611937 |
| 3 - ALL (-ANIMALS) | Component | cytoplasmic membrane-bounded vesicle | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Component | cytoskeleton | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Component | endomembrane system | (none in phylogroup) | 0.8864925 |
| 3 - ALL (-ANIMALS) | Component | endoplasmic reticulum | (none in phylogroup) | 0.772402 |
| 3 - ALL (-ANIMALS) | Component | extracellular region | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Component | Golgi apparatus | -0.587931245 | 1 |
| 3 - ALL (-ANIMALS) | Component | membrane | -1.216692299 | 0.326697 |
| 3 - ALL (-ANIMALS) | Component | membrane fraction | 0.842703109 | 1 |
| 3 - ALL (-ANIMALS) | Component | microtubule organizing center | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Component | mitochondrial envelope | 0.20976058 | 1 |
| 3 - ALL (-ANIMALS) | Component | mitochondrion | 0.396638714 | 1 |
| 3 - ALL (-ANIMALS) | Component | nucleolus | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Component | nucleus | -0.750598692 | 0.5337381 |
| 3 - ALL (-ANIMALS) | Component | peroxisome | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Component | plasma membrane | 0.361264384 | 1 |
| 3 - ALL (-ANIMALS) | Component | ribosome | (none in phylogroup) | 0.91771 |
| 3 - ALL (-ANIMALS) | Component | site of polarized growth | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Component | vacuole | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | ATPase activity | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | cytoskeletal protein binding | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | DNA binding | -1.466624949 | 1 |
| 3 - ALL (-ANIMALS) | Function | enzyme binding | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | enzyme regulator activity | -0.816956759 | 1 |
| 3 - ALL (-ANIMALS) | Function | GTPase activity | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | helicase activity | 0.629299471 | 1 |
| 3 - ALL (-ANIMALS) | Function | histone binding | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 2.020490229 | 0.2998789 |
| 3 - ALL (-ANIMALS) | Function | hydrolase activity, acting on glycosyl bonds | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | ion binding | 1.593675561 | 0.785908 |
| 3 - ALL (-ANIMALS) | Function | isomerase activity | 2.118337552 | 0.2574422 |
| 3 - ALL (-ANIMALS) | Function | kinase activity | 2.100371005 | 0.008020859 |
| 3 - ALL (-ANIMALS) | Function | ligase activity | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | lipid binding | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | lyase activity | 4.383543057 | 0 |
| 3 - ALL (-ANIMALS) | Function | methyltransferase activity | 1.524962811 | 0.6260293 |
| 3 - ALL (-ANIMALS) | Function | molecular function unknown | -3.013113302 | 7.21E-05 |
| 3 - ALL (-ANIMALS) | Function | mRNA binding | 0.703300053 | 1 |
| 3 - ALL (-ANIMALS) | Function | nuclease activity | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | nucleic acid binding transcription factor activity | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | nucleotidyltransferase activity | 0.131048604 | 1 |
| 3 - ALL (-ANIMALS) | Function | oxidoreductase activity | 2.302134728 | 9.13E-05 |
| 3 - ALL (-ANIMALS) | Function | peptidase activity | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | phosphatase activity | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | protein binding transcription factor activity | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | protein binding, bridging | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | protein transporter activity | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | RNA binding | -2.607193147 | 0.279941 |
| 3 - ALL (-ANIMALS) | Function | RNA modification guide activity | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | rRNA binding | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | signal transducer activity | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | small conjugating protein binding | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | structural constituent of ribosome | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | structural molecule activity | (none in phylogroup) | 0.9063606 |
| 3 - ALL (-ANIMALS) | Function | transcription factor binding | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | transferase activity, transferring acyl groups | 1.143872644 | 1 |
| 3 - ALL (-ANIMALS) | Function | transferase activity, transferring alkyl or aryl (other than methyl) groups | 3.665825347 | 0.000731994 |
| 3 - ALL (-ANIMALS) | Function | transferase activity, transferring glycosyl groups | 1.307371376 | 0.8339068 |
| 3 - ALL (-ANIMALS) | Function | translation factor activity, nucleic acid binding | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | transmembrane transporter activity | 0.684441025 | 1 |
| 3 - ALL (-ANIMALS) | Function | triplet codon-amino acid adaptor activity | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Function | unfolded protein binding | 0.885138376 | 1 |
| 3 - ALL (-ANIMALS) | Process | amino acid transport | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | biological process unknown | -2.29432514 | 0.04985459 |
| 3 - ALL (-ANIMALS) | Process | carbohydrate metabolic process | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | carbohydrate transport | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | cell budding | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | cell morphogenesis | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | cell wall organization or biogenesis | -0.823559494 | 1 |
| 3 - ALL (-ANIMALS) | Process | cellular amino acid metabolic process | 3.851691893 | 0 |
| 3 - ALL (-ANIMALS) | Process | cellular ion homeostasis | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | cellular respiration | 0.42766561 | 1 |
| 3 - ALL (-ANIMALS) | Process | chromatin organization | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | chromosome segregation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | cofactor metabolic process | 1.933305658 | 0.05666634 |
| 3 - ALL (-ANIMALS) | Process | conjugation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | cytokinesis | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | cytoplasmic translation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | cytoskeleton organization | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | DNA recombination | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | DNA repair | -0.894262485 | 1 |
| 3 - ALL (-ANIMALS) | Process | DNA replication | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | endocytosis | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | endosome transport | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | exocytosis | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | generation of precursor metabolites and energy | -0.415094648 | 1 |
| 3 - ALL (-ANIMALS) | Process | Golgi vesicle transport | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | histone modification | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | invasive growth in response to glucose limitation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | ion transport | 1.917804565 | 0.1122906 |
| 3 - ALL (-ANIMALS) | Process | lipid metabolic process | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | lipid transport | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | meiotic cell cycle | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | membrane fusion | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | membrane invagination | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | mitochondrial translation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | mitochondrion organization | -1.393068342 | 1 |
| 3 - ALL (-ANIMALS) | Process | mitotic cell cycle | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | mRNA processing | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | nuclear transport | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | nucleobase, nucleoside and nucleotide metabolic process | 0.42766561 | 1 |
| 3 - ALL (-ANIMALS) | Process | nucleobase, nucleoside, nucleotide and nucleic acid transport | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | nucleus organization | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | oligosaccharide metabolic process | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | organelle assembly | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | organelle fission | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | organelle fusion | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | organelle inheritance | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | peptidyl-amino acid modification | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | peroxisome organization | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | protein acylation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | protein alkylation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | protein complex biogenesis | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | protein dephosphorylation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | protein folding | 0.491795947 | 1 |
| 3 - ALL (-ANIMALS) | Process | protein glycosylation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | protein lipidation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | protein maturation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | protein modification by small protein conjugation or removal | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | protein phosphorylation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | protein targeting | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | proteolysis involved in cellular protein catabolic process | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | pseudohyphal growth | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | regulation of cell cycle | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | regulation of DNA metabolic process | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | regulation of organelle organization | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | regulation of protein modification process | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | regulation of translation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | regulation of transport | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | response to chemical stimulus | 0.131048604 | 1 |
| 3 - ALL (-ANIMALS) | Process | response to DNA damage stimulus | -1.188323786 | 1 |
| 3 - ALL (-ANIMALS) | Process | response to heat | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | response to osmotic stress | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | response to oxidative stress | 2.126799131 | 0.1316088 |
| 3 - ALL (-ANIMALS) | Process | response to starvation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | ribosomal large subunit biogenesis | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | ribosomal small subunit biogenesis | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | ribosomal subunit export from nucleus | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | ribosome assembly | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | RNA catabolic process | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | RNA modification | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | RNA splicing | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | rRNA processing | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | signaling | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | snoRNA processing | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | sporulation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | telomere organization | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | transcription elongation, DNA-dependent | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | transcription from RNA polymerase I promoter | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | transcription from RNA polymerase II promoter | (none in phylogroup) | 0.5813516 |
| 3 - ALL (-ANIMALS) | Process | transcription from RNA polymerase III promoter | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | transcription initiation, DNA-dependent | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | transcription termination, DNA-dependent | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | translational elongation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | translational initiation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | transmembrane transport | -0.603361286 | 1 |
| 3 - ALL (-ANIMALS) | Process | transposition | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | tRNA aminoacylation for protein translation | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | tRNA processing | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | vacuole organization | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | vesicle organization | (none in phylogroup) | 1 |
| 3 - ALL (-ANIMALS) | Process | vitamin metabolic process | 4.856070333 | 2.04E-13 |
| 4 - EUKARYOTES | Component | cell cortex | 0.245389562 | 1 |
| 4 - EUKARYOTES | Component | cell wall | -1.512830951 | 0.00104333 |
| 4 - EUKARYOTES | Component | cellular bud | -0.172278194 | 1 |
| 4 - EUKARYOTES | Component | cellular component unknown | -1.951949043 | 4.82E-43 |
| 4 - EUKARYOTES | Component | chromosome | 0.622470034 | 1.35E-07 |
| 4 - EUKARYOTES | Component | cytoplasm | 0.109612725 | 9.05E-05 |
| 4 - EUKARYOTES | Component | cytoplasmic membrane-bounded vesicle | 1.106427241 | 6.38E-10 |
| 4 - EUKARYOTES | Component | cytoskeleton | 0.202911372 | 1 |
| 4 - EUKARYOTES | Component | endomembrane system | 0.913110057 | 0 |
| 4 - EUKARYOTES | Component | endoplasmic reticulum | 0.636505071 | 6.42E-09 |
| 4 - EUKARYOTES | Component | extracellular region | -0.515802517 | 1 |
| 4 - EUKARYOTES | Component | Golgi apparatus | 0.947853687 | 1.96E-11 |
| 4 - EUKARYOTES | Component | membrane | 0.418830776 | 0 |
| 4 - EUKARYOTES | Component | membrane fraction | 0.246037746 | 0.8200964 |
| 4 - EUKARYOTES | Component | microtubule organizing center | 0.312294832 | 1 |
| 4 - EUKARYOTES | Component | mitochondrial envelope | 0.398082325 | 0.01016962 |
| 4 - EUKARYOTES | Component | mitochondrion | -0.124362379 | 0.6746366 |
| 4 - EUKARYOTES | Component | nucleolus | 0.702306018 | 2.14E-09 |
| 4 - EUKARYOTES | Component | nucleus | 0.438366969 | 0 |
| 4 - EUKARYOTES | Component | peroxisome | 0.354220696 | 1 |
| 4 - EUKARYOTES | Component | plasma membrane | -0.529848338 | 0.02119529 |
| 4 - EUKARYOTES | Component | ribosome | -0.313456038 | 0.2949864 |
| 4 - EUKARYOTES | Component | site of polarized growth | 0.090608403 | 1 |
| 4 - EUKARYOTES | Component | vacuole | 0.401659209 | 0.05359805 |
| 4 - EUKARYOTES | Function | ATPase activity | 0.839851164 | 1.98E-10 |
| 4 - EUKARYOTES | Function | cytoskeletal protein binding | 0.354220696 | 1 |
| 4 - EUKARYOTES | Function | DNA binding | 0.053893227 | 1 |
| 4 - EUKARYOTES | Function | enzyme binding | 0.332194389 | 1 |
| 4 - EUKARYOTES | Function | enzyme regulator activity | 0.452753313 | 0.01764873 |
| 4 - EUKARYOTES | Function | GTPase activity | 0.784706594 | 0.01977614 |
| 4 - EUKARYOTES | Function | helicase activity | 0.765153797 | 0.004720123 |
| 4 - EUKARYOTES | Function | histone binding | 0.91715689 | 0.03087782 |
| 4 - EUKARYOTES | Function | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | -0.691652353 | 0.8145655 |
| 4 - EUKARYOTES | Function | hydrolase activity, acting on glycosyl bonds | -0.637431962 | 1 |
| 4 - EUKARYOTES | Function | ion binding | 0.951922308 | 0.008103135 |
| 4 - EUKARYOTES | Function | isomerase activity | 0.171729717 | 1 |
| 4 - EUKARYOTES | Function | kinase activity | 0.633984839 | 0.000105741 |
| 4 - EUKARYOTES | Function | ligase activity | 0.379500104 | 0.1681981 |
| 4 - EUKARYOTES | Function | lipid binding | 0.68366676 | 0.0139711 |
| 4 - EUKARYOTES | Function | lyase activity | -1.28795754 | 0.01436305 |
| 4 - EUKARYOTES | Function | methyltransferase activity | 0.776294354 | 0.002252199 |
| 4 - EUKARYOTES | Function | molecular function unknown | -0.458469342 | 3.35E-17 |
| 4 - EUKARYOTES | Function | mRNA binding | 0.756692218 | 0.007545951 |
| 4 - EUKARYOTES | Function | nuclease activity | -0.198320327 | 1 |
| 4 - EUKARYOTES | Function | nucleic acid binding transcription factor activity | -1.684879124 | 0.001076143 |
| 4 - EUKARYOTES | Function | nucleotidyltransferase activity | 0.418906022 | 0.305377 |
| 4 - EUKARYOTES | Function | oxidoreductase activity | -0.490927849 | 0.05009132 |
| 4 - EUKARYOTES | Function | peptidase activity | 0.141052902 | 1 |
| 4 - EUKARYOTES | Function | phosphatase activity | 0.674796052 | 0.009371203 |
| 4 - EUKARYOTES | Function | protein binding transcription factor activity | -0.523415701 | 1 |
| 4 - EUKARYOTES | Function | protein binding, bridging | 0.817621216 | 0.03501744 |
| 4 - EUKARYOTES | Function | protein transporter activity | 1.05208647 | 0.000244975 |
| 4 - EUKARYOTES | Function | RNA binding | -0.522322751 | 2.26E-06 |
| 4 - EUKARYOTES | Function | RNA modification guide activity | (none in phylogroup) | 3.21E-08 |
| 4 - EUKARYOTES | Function | rRNA binding | -1.162570302 | 0.01616457 |
| 4 - EUKARYOTES | Function | signal transducer activity | -0.597416283 | 1 |
| 4 - EUKARYOTES | Function | small conjugating protein binding | 1.014454091 | 0.002037254 |
| 4 - EUKARYOTES | Function | structural constituent of ribosome | -0.976852414 | 9.13E-05 |
| 4 - EUKARYOTES | Function | structural molecule activity | -0.302521147 | 0.338017 |
| 4 - EUKARYOTES | Function | transcription factor binding | -0.523415701 | 1 |
| 4 - EUKARYOTES | Function | transferase activity, transferring acyl groups | 0.633363924 | 0.009879267 |
| 4 - EUKARYOTES | Function | transferase activity, transferring alkyl or aryl (other than methyl) groups | -0.368245329 | 1 |
| 4 - EUKARYOTES | Function | transferase activity, transferring glycosyl groups | -0.203137344 | 1 |
| 4 - EUKARYOTES | Function | translation factor activity, nucleic acid binding | 0.641522447 | 0.2260198 |
| 4 - EUKARYOTES | Function | transmembrane transporter activity | 0.12610378 | 1 |
| 4 - EUKARYOTES | Function | triplet codon-amino acid adaptor activity | (none in phylogroup) | 2.23E-39 |
| 4 - EUKARYOTES | Function | unfolded protein binding | 0.709048695 | 0.03067461 |
| 4 - EUKARYOTES | Process | amino acid transport | -0.957312228 | 0.6400706 |
| 4 - EUKARYOTES | Process | biological process unknown | -1.362611532 | 1.54E-50 |
| 4 - EUKARYOTES | Process | carbohydrate metabolic process | -0.226678883 | 1 |
| 4 - EUKARYOTES | Process | carbohydrate transport | -0.330770623 | 1 |
| 4 - EUKARYOTES | Process | cell budding | 0.262653456 | 1 |
| 4 - EUKARYOTES | Process | cell morphogenesis | 0.029631619 | 1 |
| 4 - EUKARYOTES | Process | cell wall organization or biogenesis | -0.648176804 | 0.0165155 |
| 4 - EUKARYOTES | Process | cellular amino acid metabolic process | -1.345877516 | 3.50E-08 |
| 4 - EUKARYOTES | Process | cellular ion homeostasis | 0.222569898 | 1 |
| 4 - EUKARYOTES | Process | cellular respiration | 0.393594934 | 0.5864367 |
| 4 - EUKARYOTES | Process | chromatin organization | 0.763682409 | 7.73E-08 |
| 4 - EUKARYOTES | Process | chromosome segregation | 0.595228795 | 0.00808681 |
| 4 - EUKARYOTES | Process | cofactor metabolic process | -0.422693113 | 0.5702022 |
| 4 - EUKARYOTES | Process | conjugation | 0.04679217 | 1 |
| 4 - EUKARYOTES | Process | cytokinesis | -0.056847901 | 1 |
| 4 - EUKARYOTES | Process | cytoplasmic translation | -0.099521851 | 1 |
| 4 - EUKARYOTES | Process | cytoskeleton organization | 0.604987762 | 0.000126968 |
| 4 - EUKARYOTES | Process | DNA recombination | 0.498844259 | 0.04019729 |
| 4 - EUKARYOTES | Process | DNA repair | 1.048946763 | 0 |
| 4 - EUKARYOTES | Process | DNA replication | 0.762828744 | 5.43E-05 |
| 4 - EUKARYOTES | Process | endocytosis | 0.689746394 | 0.007458264 |
| 4 - EUKARYOTES | Process | endosome transport | 0.936265713 | 0.000129131 |
| 4 - EUKARYOTES | Process | exocytosis | 0.821999657 | 0.02728584 |
| 4 - EUKARYOTES | Process | generation of precursor metabolites and energy | -0.02290057 | 1 |
| 4 - EUKARYOTES | Process | Golgi vesicle transport | 1.129377506 | 0 |
| 4 - EUKARYOTES | Process | histone modification | 1.004619731 | 6.34E-07 |
| 4 - EUKARYOTES | Process | invasive growth in response to glucose limitation | 0.332194389 | 1 |
| 4 - EUKARYOTES | Process | ion transport | -0.308911189 | 1 |
| 4 - EUKARYOTES | Process | lipid metabolic process | 0.802597126 | 1.22E-10 |
| 4 - EUKARYOTES | Process | lipid transport | 0.624375141 | 0.2291574 |
| 4 - EUKARYOTES | Process | meiotic cell cycle | 0.545708659 | 0.008746135 |
| 4 - EUKARYOTES | Process | membrane fusion | 0.989306676 | 1.83E-05 |
| 4 - EUKARYOTES | Process | membrane invagination | 0.712337098 | 0.000320789 |
| 4 - EUKARYOTES | Process | mitochondrial translation | -2.574696206 | 1.60E-09 |
| 4 - EUKARYOTES | Process | mitochondrion organization | -0.179211504 | 1 |
| 4 - EUKARYOTES | Process | mitotic cell cycle | 0.65005282 | 5.90E-07 |
| 4 - EUKARYOTES | Process | mRNA processing | 1.174808099 | 0 |
| 4 - EUKARYOTES | Process | nuclear transport | 0.953226145 | 7.45E-10 |
| 4 - EUKARYOTES | Process | nucleobase, nucleoside and nucleotide metabolic process | -0.962548876 | 0.000778713 |
| 4 - EUKARYOTES | Process | nucleobase, nucleoside, nucleotide and nucleic acid transport | 0.510531631 | 0.08460201 |
| 4 - EUKARYOTES | Process | nucleus organization | 0.728123066 | 0.04534108 |
| 4 - EUKARYOTES | Process | oligosaccharide metabolic process | 0.408143243 | 1 |
| 4 - EUKARYOTES | Process | organelle assembly | 1.144567386 | 2.08E-08 |
| 4 - EUKARYOTES | Process | organelle fission | 0.824047486 | 1.45E-05 |
| 4 - EUKARYOTES | Process | organelle fusion | 1.194690866 | 1.13E-07 |
| 4 - EUKARYOTES | Process | organelle inheritance | 0.860573362 | 0.009624287 |
| 4 - EUKARYOTES | Process | peptidyl-amino acid modification | 0.991157471 | 2.31E-05 |
| 4 - EUKARYOTES | Process | peroxisome organization | 0.353256005 | 1 |
| 4 - EUKARYOTES | Process | protein acylation | 1.225279185 | 7.17E-08 |
| 4 - EUKARYOTES | Process | protein alkylation | 1.180191296 | 0.000113766 |
| 4 - EUKARYOTES | Process | protein complex biogenesis | 0.82630346 | 2.80E-09 |
| 4 - EUKARYOTES | Process | protein dephosphorylation | 0.756692218 | 0.1253657 |
| 4 - EUKARYOTES | Process | protein folding | 0.981287227 | 5.34E-06 |
| 4 - EUKARYOTES | Process | protein glycosylation | 0.509072151 | 0.3252427 |
| 4 - EUKARYOTES | Process | protein lipidation | 1.490892135 | 1.04E-09 |
| 4 - EUKARYOTES | Process | protein maturation | 0.538645267 | 0.6552095 |
| 4 - EUKARYOTES | Process | protein modification by small protein conjugation or removal | 1.146638736 | 0 |
| 4 - EUKARYOTES | Process | protein phosphorylation | 0.848677152 | 5.02E-07 |
| 4 - EUKARYOTES | Process | protein targeting | 0.950071512 | 0 |
| 4 - EUKARYOTES | Process | proteolysis involved in cellular protein catabolic process | 1.005512764 | 2.04E-13 |
| 4 - EUKARYOTES | Process | pseudohyphal growth | -0.1489323 | 1 |
| 4 - EUKARYOTES | Process | regulation of cell cycle | 0.674043178 | 5.18E-05 |
| 4 - EUKARYOTES | Process | regulation of DNA metabolic process | 0.284888674 | 1 |
| 4 - EUKARYOTES | Process | regulation of organelle organization | 0.790624484 | 3.67E-05 |
| 4 - EUKARYOTES | Process | regulation of protein modification process | 0.747231889 | 0.0223337 |
| 4 - EUKARYOTES | Process | regulation of translation | 0.332194389 | 0.9809182 |
| 4 - EUKARYOTES | Process | regulation of transport | 0.784706594 | 0.01977614 |
| 4 - EUKARYOTES | Process | response to chemical stimulus | -0.221198032 | 0.972524 |
| 4 - EUKARYOTES | Process | response to DNA damage stimulus | 0.977277882 | 0 |
| 4 - EUKARYOTES | Process | response to heat | -0.046317234 | 1 |
| 4 - EUKARYOTES | Process | response to osmotic stress | -0.272667669 | 1 |
| 4 - EUKARYOTES | Process | response to oxidative stress | -0.492234046 | 1 |
| 4 - EUKARYOTES | Process | response to starvation | 0.732732319 | 0.01227145 |
| 4 - EUKARYOTES | Process | ribosomal large subunit biogenesis | 1.144567386 | 2.08E-08 |
| 4 - EUKARYOTES | Process | ribosomal small subunit biogenesis | 0.795850594 | 5.51E-05 |
| 4 - EUKARYOTES | Process | ribosomal subunit export from nucleus | 1.225279185 | 2.23E-05 |
| 4 - EUKARYOTES | Process | ribosome assembly | 1.034513841 | 9.95E-05 |
| 4 - EUKARYOTES | Process | RNA catabolic process | 0.930577406 | 2.56E-06 |
| 4 - EUKARYOTES | Process | RNA modification | -0.064227432 | 1 |
| 4 - EUKARYOTES | Process | RNA splicing | 1.06705232 | 1.72E-11 |
| 4 - EUKARYOTES | Process | rRNA processing | 0.597697051 | 6.79E-06 |
| 4 - EUKARYOTES | Process | signaling | 0.094034652 | 1 |
| 4 - EUKARYOTES | Process | snoRNA processing | 0.637048971 | 0.4272734 |
| 4 - EUKARYOTES | Process | sporulation | 0.320789626 | 0.7140367 |
| 4 - EUKARYOTES | Process | telomere organization | 0.627650273 | 0.1000495 |
| 4 - EUKARYOTES | Process | transcription elongation, DNA-dependent | 1.035801386 | 1.54E-05 |
| 4 - EUKARYOTES | Process | transcription from RNA polymerase I promoter | 0.991157471 | 0.00038039 |
| 4 - EUKARYOTES | Process | transcription from RNA polymerase II promoter | 0.297760892 | 0.05283757 |
| 4 - EUKARYOTES | Process | transcription from RNA polymerase III promoter | 0.91715689 | 0.01497375 |
| 4 - EUKARYOTES | Process | transcription initiation, DNA-dependent | 0.956685254 | 0.000111729 |
| 4 - EUKARYOTES | Process | transcription termination, DNA-dependent | 1.097729136 | 0.004933033 |
| 4 - EUKARYOTES | Process | translational elongation | -3.453530517 | 1.10E-31 |
| 4 - EUKARYOTES | Process | translational initiation | 0.633363924 | 0.1507798 |
| 4 - EUKARYOTES | Process | transmembrane transport | 0.143927752 | 1 |
| 4 - EUKARYOTES | Process | transposition | -1.837730612 | 3.04E-05 |
| 4 - EUKARYOTES | Process | tRNA aminoacylation for protein translation | -3.292296476 | 0.007008556 |
| 4 - EUKARYOTES | Process | tRNA processing | 0.65999905 | 0.01184699 |
| 4 - EUKARYOTES | Process | vacuole organization | 0.752770072 | 0.000136695 |
| 4 - EUKARYOTES | Process | vesicle organization | 1.378290804 | 2.04E-13 |
| 4 - EUKARYOTES | Process | vitamin metabolic process | -2.637431962 | 0.003713731 |
| 5 - ANIMALS + FUNGI | Component | cell cortex | 2.16806882 | 2.22E-07 |
| 5 - ANIMALS + FUNGI | Component | cell wall | -1.655740034 | 1 |
| 5 - ANIMALS + FUNGI | Component | cellular bud | 1.128912688 | 0.07215885 |
| 5 - ANIMALS + FUNGI | Component | cellular component unknown | -1.08021135 | 0.06766475 |
| 5 - ANIMALS + FUNGI | Component | chromosome | -0.299530119 | 1 |
| 5 - ANIMALS + FUNGI | Component | cytoplasm | 0.25729377 | 0.005631861 |
| 5 - ANIMALS + FUNGI | Component | cytoplasmic membrane-bounded vesicle | 1.202705822 | 0.1953396 |
| 5 - ANIMALS + FUNGI | Component | cytoskeleton | 1.551855386 | 0.000225876 |
| 5 - ANIMALS + FUNGI | Component | endomembrane system | 0.271747467 | 1 |
| 5 - ANIMALS + FUNGI | Component | endoplasmic reticulum | 0.700046865 | 0.2633169 |
| 5 - ANIMALS + FUNGI | Component | extracellular region | (none in phylogroup) | 1 |
| 5 - ANIMALS + FUNGI | Component | Golgi apparatus | 0.864445616 | 0.3911941 |
| 5 - ANIMALS + FUNGI | Component | membrane | 0.683143539 | 0.000122728 |
| 5 - ANIMALS + FUNGI | Component | membrane fraction | 0.71011747 | 0.6918986 |
| 5 - ANIMALS + FUNGI | Component | microtubule organizing center | -0.24565175 | 1 |
| 5 - ANIMALS + FUNGI | Component | mitochondrial envelope | 0.31818304 | 1 |
| 5 - ANIMALS + FUNGI | Component | mitochondrion | 0.230886211 | 1 |
| 5 - ANIMALS + FUNGI | Component | nucleolus | -1.368710147 | 0.4306717 |
| 5 - ANIMALS + FUNGI | Component | nucleus | -0.379141827 | 0.2425508 |
| 5 - ANIMALS + FUNGI | Component | peroxisome | 1.506767496 | 0.132819 |
| 5 - ANIMALS + FUNGI | Component | plasma membrane | 0.769247126 | 0.2921 |
| 5 - ANIMALS + FUNGI | Component | ribosome | -0.930296309 | 0.9328225 |
| 5 - ANIMALS + FUNGI | Component | site of polarized growth | 1.439552416 | 0.000699454 |
| 5 - ANIMALS + FUNGI | Component | vacuole | 0.843712627 | 0.3089402 |
| 5 - ANIMALS + FUNGI | Function | ATPase activity | -0.579389148 | 1 |
| 5 - ANIMALS + FUNGI | Function | cytoskeletal protein binding | 2.24373309 | 0.000363075 |
| 5 - ANIMALS + FUNGI | Function | DNA binding | 0.111282794 | 1 |
| 5 - ANIMALS + FUNGI | Function | enzyme binding | 0.65877059 | 1 |
| 5 - ANIMALS + FUNGI | Function | enzyme regulator activity | 0.175988484 | 1 |
| 5 - ANIMALS + FUNGI | Function | GTPase activity | 1.696245295 | 0.06944775 |
| 5 - ANIMALS + FUNGI | Function | helicase activity | (none in phylogroup) | 1 |
| 5 - ANIMALS + FUNGI | Function | histone binding | 1.441672468 | 0.4949317 |
| 5 - ANIMALS + FUNGI | Function | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 0.598397972 | 1 |
| 5 - ANIMALS + FUNGI | Function | hydrolase activity, acting on glycosyl bonds | 0.389583957 | 1 |
| 5 - ANIMALS + FUNGI | Function | ion binding | (none in phylogroup) | 1 |
| 5 - ANIMALS + FUNGI | Function | isomerase activity | 0.111282794 | 1 |
| 5 - ANIMALS + FUNGI | Function | kinase activity | 1.678278749 | 3.68E-05 |
| 5 - ANIMALS + FUNGI | Function | ligase activity | 0.259674634 | 1 |
| 5 - ANIMALS + FUNGI | Function | lipid binding | 1.501229313 | 0.06000298 |
| 5 - ANIMALS + FUNGI | Function | lyase activity | 0.154095878 | 1 |
| 5 - ANIMALS + FUNGI | Function | methyltransferase activity | 0.517908054 | 1 |
| 5 - ANIMALS + FUNGI | Function | molecular function unknown | -0.593903305 | 0.0143135 |
| 5 - ANIMALS + FUNGI | Function | mRNA binding | -0.303754705 | 1 |
| 5 - ANIMALS + FUNGI | Function | nuclease activity | -0.493232504 | 1 |
| 5 - ANIMALS + FUNGI | Function | nucleic acid binding transcription factor activity | 0.119744373 | 1 |
| 5 - ANIMALS + FUNGI | Function | nucleotidyltransferase activity | -0.876006154 | 1 |
| 5 - ANIMALS + FUNGI | Function | oxidoreductase activity | -1.164351648 | 0.8801834 |
| 5 - ANIMALS + FUNGI | Function | peptidase activity | 0.431103226 | 1 |
| 5 - ANIMALS + FUNGI | Function | phosphatase activity | 0.666188061 | 1 |
| 5 - ANIMALS + FUNGI | Function | protein binding transcription factor activity | -0.303754705 | 1 |
| 5 - ANIMALS + FUNGI | Function | protein binding, bridging | 1.037282213 | 0.9581992 |
| 5 - ANIMALS + FUNGI | Function | protein transporter activity | 1.271747467 | 0.5058428 |
| 5 - ANIMALS + FUNGI | Function | RNA binding | -2.29231981 | 4.16E-05 |
| 5 - ANIMALS + FUNGI | Function | RNA modification guide activity | (none in phylogroup) | 1 |
| 5 - ANIMALS + FUNGI | Function | rRNA binding | (none in phylogroup) | 0.8200964 |
| 5 - ANIMALS + FUNGI | Function | signal transducer activity | 2.429599636 | 0.001695868 |
| 5 - ANIMALS + FUNGI | Function | small conjugating protein binding | 1.839836149 | 0.07300097 |
| 5 - ANIMALS + FUNGI | Function | structural constituent of ribosome | -2.856727091 | 0.132819 |
| 5 - ANIMALS + FUNGI | Function | structural molecule activity | -1.201504647 | 0.4882773 |
| 5 - ANIMALS + FUNGI | Function | transcription factor binding | (none in phylogroup) | 1 |
| 5 - ANIMALS + FUNGI | Function | transferase activity, transferring acyl groups | 0.136817886 | 1 |
| 5 - ANIMALS + FUNGI | Function | transferase activity, transferring alkyl or aryl (other than methyl) groups | -0.34122941 | 1 |
| 5 - ANIMALS + FUNGI | Function | transferase activity, transferring glycosyl groups | 1.47024162 | 0.04654288 |
| 5 - ANIMALS + FUNGI | Function | translation factor activity, nucleic acid binding | -0.579389148 | 1 |
| 5 - ANIMALS + FUNGI | Function | transmembrane transporter activity | 0.262348769 | 1 |
| 5 - ANIMALS + FUNGI | Function | triplet codon-amino acid adaptor activity | (none in phylogroup) | 0.001975812 |
| 5 - ANIMALS + FUNGI | Function | unfolded protein binding | -1.121916382 | 1 |
| 5 - ANIMALS + FUNGI | Process | amino acid transport | 1.48474119 | 0.3084608 |
| 5 - ANIMALS + FUNGI | Process | biological process unknown | -0.657523708 | 0.1241817 |
| 5 - ANIMALS + FUNGI | Process | carbohydrate metabolic process | 0.653495648 | 0.6370665 |
| 5 - ANIMALS + FUNGI | Process | carbohydrate transport | -0.303754705 | 1 |
| 5 - ANIMALS + FUNGI | Process | cell budding | 2.119744373 | 0.000179097 |
| 5 - ANIMALS + FUNGI | Process | cell morphogenesis | 1.734719443 | 0.1676199 |
| 5 - ANIMALS + FUNGI | Process | cell wall organization or biogenesis | 0.491313844 | 1 |
| 5 - ANIMALS + FUNGI | Process | cellular amino acid metabolic process | -0.640789692 | 1 |
| 5 - ANIMALS + FUNGI | Process | cellular ion homeostasis | 0.001658303 | 1 |
| 5 - ANIMALS + FUNGI | Process | cellular respiration | -0.579389148 | 1 |
| 5 - ANIMALS + FUNGI | Process | chromatin organization | 0.876428202 | 0.2661428 |
| 5 - ANIMALS + FUNGI | Process | chromosome segregation | -0.132642789 | 1 |
| 5 - ANIMALS + FUNGI | Process | cofactor metabolic process | -0.073749099 | 1 |
| 5 - ANIMALS + FUNGI | Process | conjugation | 0.881163011 | 0.6850835 |
| 5 - ANIMALS + FUNGI | Process | cytokinesis | 1.747775596 | 0.002428423 |
| 5 - ANIMALS + FUNGI | Process | cytoplasmic translation | -2.498770687 | 0.4479066 |
| 5 - ANIMALS + FUNGI | Process | cytoskeleton organization | 1.615701868 | 4.41E-05 |
| 5 - ANIMALS + FUNGI | Process | DNA recombination | -0.255499536 | 1 |
| 5 - ANIMALS + FUNGI | Process | DNA repair | -0.09396232 | 1 |
| 5 - ANIMALS + FUNGI | Process | DNA replication | 0.157576447 | 1 |
| 5 - ANIMALS + FUNGI | Process | endocytosis | 1.818641926 | 0.002710594 |
| 5 - ANIMALS + FUNGI | Process | endosome transport | 2.174785737 | 0.000260595 |
| 5 - ANIMALS + FUNGI | Process | exocytosis | 2.711512052 | 1.63E-05 |
| 5 - ANIMALS + FUNGI | Process | generation of precursor metabolites and energy | 0.747775596 | 0.772402 |
| 5 - ANIMALS + FUNGI | Process | Golgi vesicle transport | 1.598397972 | 0.000364916 |
| 5 - ANIMALS + FUNGI | Process | histone modification | 0.681138403 | 1 |
| 5 - ANIMALS + FUNGI | Process | invasive growth in response to glucose limitation | 1.271747467 | 0.5058428 |
| 5 - ANIMALS + FUNGI | Process | ion transport | 0.495712308 | 1 |
| 5 - ANIMALS + FUNGI | Process | lipid metabolic process | 0.910749807 | 0.132819 |
| 5 - ANIMALS + FUNGI | Process | lipid transport | 1.651391059 | 0.1318774 |
| 5 - ANIMALS + FUNGI | Process | meiotic cell cycle | -0.082627251 | 1 |
| 5 - ANIMALS + FUNGI | Process | membrane fusion | 0.24373309 | 1 |
| 5 - ANIMALS + FUNGI | Process | membrane invagination | 1.557149685 | 0.005423144 |
| 5 - ANIMALS + FUNGI | Process | mitochondrial translation | -2.132642789 | 1 |
| 5 - ANIMALS + FUNGI | Process | mitochondrion organization | -0.078195004 | 1 |
| 5 - ANIMALS + FUNGI | Process | mitotic cell cycle | 0.116353784 | 1 |
| 5 - ANIMALS + FUNGI | Process | mRNA processing | -0.539642969 | 1 |
| 5 - ANIMALS + FUNGI | Process | nuclear transport | -0.35960794 | 1 |
| 5 - ANIMALS + FUNGI | Process | nucleobase, nucleoside and nucleotide metabolic process | 0.590535854 | 1 |
| 5 - ANIMALS + FUNGI | Process | nucleobase, nucleoside, nucleotide and nucleic acid transport | 0.696245295 | 1 |
| 5 - ANIMALS + FUNGI | Process | nucleus organization | -0.888717206 | 1 |
| 5 - ANIMALS + FUNGI | Process | oligosaccharide metabolic process | -0.265280557 | 1 |
| 5 - ANIMALS + FUNGI | Process | organelle assembly | -1.413379196 | 1 |
| 5 - ANIMALS + FUNGI | Process | organelle fission | -2.055827191 | 1 |
| 5 - ANIMALS + FUNGI | Process | organelle fusion | -1.100221311 | 1 |
| 5 - ANIMALS + FUNGI | Process | organelle inheritance | 1.565661185 | 0.1669297 |
| 5 - ANIMALS + FUNGI | Process | peptidyl-amino acid modification | -0.303754705 | 1 |
| 5 - ANIMALS + FUNGI | Process | peroxisome organization | -0.143290033 | 1 |
| 5 - ANIMALS + FUNGI | Process | protein acylation | -1.033107115 | 1 |
| 5 - ANIMALS + FUNGI | Process | protein alkylation | 1.207207214 | 0.7403704 |
| 5 - ANIMALS + FUNGI | Process | protein complex biogenesis | 0.209463188 | 1 |
| 5 - ANIMALS + FUNGI | Process | protein dephosphorylation | -0.303754705 | 1 |
| 5 - ANIMALS + FUNGI | Process | protein folding | (none in phylogroup) | 0.9232297 |
| 5 - ANIMALS + FUNGI | Process | protein glycosylation | 0.835648352 | 1 |
| 5 - ANIMALS + FUNGI | Process | protein lipidation | -0.482091946 | 1 |
| 5 - ANIMALS + FUNGI | Process | protein maturation | 0.65877059 | 1 |
| 5 - ANIMALS + FUNGI | Process | protein modification by small protein conjugation or removal | 0.349226219 | 1 |
| 5 - ANIMALS + FUNGI | Process | protein phosphorylation | 1.705768069 | 0.00038039 |
| 5 - ANIMALS + FUNGI | Process | protein targeting | 0.24373309 | 1 |
| 5 - ANIMALS + FUNGI | Process | proteolysis involved in cellular protein catabolic process | 0.544242202 | 1 |
| 5 - ANIMALS + FUNGI | Process | pseudohyphal growth | 1.878083618 | 0.01129198 |
| 5 - ANIMALS + FUNGI | Process | regulation of cell cycle | 0.719206444 | 0.7515489 |
| 5 - ANIMALS + FUNGI | Process | regulation of DNA metabolic process | 0.989976498 | 0.8809765 |
| 5 - ANIMALS + FUNGI | Process | regulation of organelle organization | 1.080674808 | 0.2415658 |
| 5 - ANIMALS + FUNGI | Process | regulation of protein modification process | 0.551855386 | 1 |
| 5 - ANIMALS + FUNGI | Process | regulation of translation | (none in phylogroup) | 0.9389088 |
| 5 - ANIMALS + FUNGI | Process | regulation of transport | 1.433210889 | 0.2420834 |
| 5 - ANIMALS + FUNGI | Process | response to chemical stimulus | 0.998462964 | 0.02115982 |
| 5 - ANIMALS + FUNGI | Process | response to DNA damage stimulus | -0.195378544 | 1 |
| 5 - ANIMALS + FUNGI | Process | response to heat | 0.65877059 | 1 |
| 5 - ANIMALS + FUNGI | Process | response to osmotic stress | 1.75434825 | 0.01977614 |
| 5 - ANIMALS + FUNGI | Process | response to oxidative stress | -1.465218128 | 1 |
| 5 - ANIMALS + FUNGI | Process | response to starvation | -0.284645882 | 1 |
| 5 - ANIMALS + FUNGI | Process | ribosomal large subunit biogenesis | -1.413379196 | 1 |
| 5 - ANIMALS + FUNGI | Process | ribosomal small subunit biogenesis | (none in phylogroup) | 0.358707 |
| 5 - ANIMALS + FUNGI | Process | ribosomal subunit export from nucleus | 1.136817886 | 0.8316331 |
| 5 - ANIMALS + FUNGI | Process | ribosome assembly | -0.938470241 | 1 |
| 5 - ANIMALS + FUNGI | Process | RNA catabolic process | -1.797294178 | 1 |
| 5 - ANIMALS + FUNGI | Process | RNA modification | -1.322613732 | 1 |
| 5 - ANIMALS + FUNGI | Process | RNA splicing | -1.153859274 | 1 |
| 5 - ANIMALS + FUNGI | Process | rRNA processing | -1.66068925 | 0.2911908 |
| 5 - ANIMALS + FUNGI | Process | signaling | 1.683645258 | 6.79E-06 |
| 5 - ANIMALS + FUNGI | Process | snoRNA processing | 0.856709967 | 1 |
| 5 - ANIMALS + FUNGI | Process | sporulation | 0.277416217 | 1 |
| 5 - ANIMALS + FUNGI | Process | telomere organization | -0.100221311 | 1 |
| 5 - ANIMALS + FUNGI | Process | transcription elongation, DNA-dependent | 1.136817886 | 0.5322395 |
| 5 - ANIMALS + FUNGI | Process | transcription from RNA polymerase I promoter | -0.888717206 | 1 |
| 5 - ANIMALS + FUNGI | Process | transcription from RNA polymerase II promoter | 0.608569777 | 0.4152864 |
| 5 - ANIMALS + FUNGI | Process | transcription from RNA polymerase III promoter | (none in phylogroup) | 1 |
| 5 - ANIMALS + FUNGI | Process | transcription initiation, DNA-dependent | (none in phylogroup) | 1 |
| 5 - ANIMALS + FUNGI | Process | transcription termination, DNA-dependent | 1.037282213 | 1 |
| 5 - ANIMALS + FUNGI | Process | translational elongation | (none in phylogroup) | 0.000731994 |
| 5 - ANIMALS + FUNGI | Process | translational initiation | -0.863182114 | 1 |
| 5 - ANIMALS + FUNGI | Process | transmembrane transport | 0.711512052 | 0.772402 |
| 5 - ANIMALS + FUNGI | Process | transposition | (none in phylogroup) | 0.5668001 |
| 5 - ANIMALS + FUNGI | Process | tRNA aminoacylation for protein translation | (none in phylogroup) | 1 |
| 5 - ANIMALS + FUNGI | Process | tRNA processing | 0.329462964 | 1 |
| 5 - ANIMALS + FUNGI | Process | vacuole organization | 1.828695591 | 0.000241313 |
| 5 - ANIMALS + FUNGI | Process | vesicle organization | -0.322613732 | 1 |
| 5 - ANIMALS + FUNGI | Process | vitamin metabolic process | -0.610416043 | 1 |
| 6 - PLANTS + FUNGI | Component | cell cortex | 0.021000433 | 1 |
| 6 - PLANTS + FUNGI | Component | cell wall | 1.519119673 | 0.02383329 |
| 6 - PLANTS + FUNGI | Component | cellular bud | -1.718595418 | 0.6858858 |
| 6 - PLANTS + FUNGI | Component | cellular component unknown | -0.364783262 | 1 |
| 6 - PLANTS + FUNGI | Component | chromosome | -2.616523508 | 0.01108979 |
| 6 - PLANTS + FUNGI | Component | cytoplasm | -0.033138793 | 1 |
| 6 - PLANTS + FUNGI | Component | cytoplasmic membrane-bounded vesicle | 0.377565529 | 1 |
| 6 - PLANTS + FUNGI | Component | cytoskeleton | -0.595213002 | 1 |
| 6 - PLANTS + FUNGI | Component | endomembrane system | -0.682675843 | 1 |
| 6 - PLANTS + FUNGI | Component | endoplasmic reticulum | 0.137940978 | 1 |
| 6 - PLANTS + FUNGI | Component | extracellular region | 2.042216919 | 0.07300097 |
| 6 - PLANTS + FUNGI | Component | Golgi apparatus | 0.427870611 | 1 |
| 6 - PLANTS + FUNGI | Component | membrane | 0.733521215 | 5.15E-06 |
| 6 - PLANTS + FUNGI | Component | membrane fraction | 1.351544977 | 0.00230258 |
| 6 - PLANTS + FUNGI | Component | microtubule organizing center | -1.392720138 | 1 |
| 6 - PLANTS + FUNGI | Component | mitochondrial envelope | -0.069893448 | 1 |
| 6 - PLANTS + FUNGI | Component | mitochondrion | 0.083817823 | 1 |
| 6 - PLANTS + FUNGI | Component | nucleolus | -0.708423612 | 1 |
| 6 - PLANTS + FUNGI | Component | nucleus | -0.672061081 | 0.001170407 |
| 6 - PLANTS + FUNGI | Component | peroxisome | -0.225263392 | 1 |
| 6 - PLANTS + FUNGI | Component | plasma membrane | 1.831632104 | 2.02E-10 |
| 6 - PLANTS + FUNGI | Component | ribosome | -0.662327198 | 1 |
| 6 - PLANTS + FUNGI | Component | site of polarized growth | -1.444481566 | 0.6485 |
| 6 - PLANTS + FUNGI | Component | vacuole | 0.318132616 | 1 |
| 6 - PLANTS + FUNGI | Function | ATPase activity | 0.652054088 | 0.697685 |
| 6 - PLANTS + FUNGI | Function | cytoskeletal protein binding | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Function | DNA binding | -0.813393172 | 1 |
| 6 - PLANTS + FUNGI | Function | enzyme binding | -0.488297798 | 1 |
| 6 - PLANTS + FUNGI | Function | enzyme regulator activity | -1.971079904 | 0.305377 |
| 6 - PLANTS + FUNGI | Function | GTPase activity | -0.035785593 | 1 |
| 6 - PLANTS + FUNGI | Function | helicase activity | -0.524823674 | 1 |
| 6 - PLANTS + FUNGI | Function | histone binding | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Function | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 0.866367083 | 1 |
| 6 - PLANTS + FUNGI | Function | hydrolase activity, acting on glycosyl bonds | 2.242515569 | 0.001765446 |
| 6 - PLANTS + FUNGI | Function | ion binding | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Function | isomerase activity | -0.035785593 | 1 |
| 6 - PLANTS + FUNGI | Function | kinase activity | 0.138892938 | 1 |
| 6 - PLANTS + FUNGI | Function | ligase activity | -0.372820581 | 1 |
| 6 - PLANTS + FUNGI | Function | lipid binding | (none in phylogroup) | 0.7504537 |
| 6 - PLANTS + FUNGI | Function | lyase activity | 0.00702749 | 1 |
| 6 - PLANTS + FUNGI | Function | methyltransferase activity | 1.178194588 | 0.2796489 |
| 6 - PLANTS + FUNGI | Function | molecular function unknown | -0.05871199 | 1 |
| 6 - PLANTS + FUNGI | Function | mRNA binding | -0.450823093 | 1 |
| 6 - PLANTS + FUNGI | Function | nuclease activity | -1.225263392 | 1 |
| 6 - PLANTS + FUNGI | Function | nucleic acid binding transcription factor activity | -0.027324015 | 1 |
| 6 - PLANTS + FUNGI | Function | nucleotidyltransferase activity | (none in phylogroup) | 0.3449095 |
| 6 - PLANTS + FUNGI | Function | oxidoreductase activity | 0.010508059 | 1 |
| 6 - PLANTS + FUNGI | Function | peptidase activity | 0.021000433 | 1 |
| 6 - PLANTS + FUNGI | Function | phosphatase activity | 1.656623197 | 0.007279627 |
| 6 - PLANTS + FUNGI | Function | protein binding transcription factor activity | -0.450823093 | 1 |
| 6 - PLANTS + FUNGI | Function | protein binding, bridging | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Function | protein transporter activity | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Function | RNA binding | -1.591391291 | 0.000985097 |
| 6 - PLANTS + FUNGI | Function | RNA modification guide activity | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Function | rRNA binding | (none in phylogroup) | 0.6344878 |
| 6 - PLANTS + FUNGI | Function | signal transducer activity | 1.060138827 | 0.931117 |
| 6 - PLANTS + FUNGI | Function | small conjugating protein binding | -0.629160334 | 1 |
| 6 - PLANTS + FUNGI | Function | structural constituent of ribosome | -1.418832978 | 0.6956062 |
| 6 - PLANTS + FUNGI | Function | structural molecule activity | -1.348573034 | 0.2423547 |
| 6 - PLANTS + FUNGI | Function | transcription factor binding | -0.450823093 | 1 |
| 6 - PLANTS + FUNGI | Function | transferase activity, transferring acyl groups | 0.574711999 | 1 |
| 6 - PLANTS + FUNGI | Function | transferase activity, transferring alkyl or aryl (other than methyl) groups | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Function | transferase activity, transferring glycosyl groups | 1.960603153 | 0.000144263 |
| 6 - PLANTS + FUNGI | Function | translation factor activity, nucleic acid binding | 0.273542465 | 1 |
| 6 - PLANTS + FUNGI | Function | transmembrane transporter activity | 1.53031788 | 1.05E-06 |
| 6 - PLANTS + FUNGI | Function | triplet codon-amino acid adaptor activity | (none in phylogroup) | 0.00073293 |
| 6 - PLANTS + FUNGI | Function | unfolded protein binding | -1.26898477 | 1 |
| 6 - PLANTS + FUNGI | Process | amino acid transport | -0.662327198 | 1 |
| 6 - PLANTS + FUNGI | Process | biological process unknown | -0.163046067 | 1 |
| 6 - PLANTS + FUNGI | Process | carbohydrate metabolic process | 0.868997339 | 0.1241817 |
| 6 - PLANTS + FUNGI | Process | carbohydrate transport | 3.636639749 | 0 |
| 6 - PLANTS + FUNGI | Process | cell budding | -1.612286515 | 1 |
| 6 - PLANTS + FUNGI | Process | cell morphogenesis | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | cell wall organization or biogenesis | 1.109780202 | 0.02930007 |
| 6 - PLANTS + FUNGI | Process | cellular amino acid metabolic process | 0.590653543 | 0.8809765 |
| 6 - PLANTS + FUNGI | Process | cellular ion homeostasis | 0.17651801 | 1 |
| 6 - PLANTS + FUNGI | Process | cellular respiration | -0.726457535 | 1 |
| 6 - PLANTS + FUNGI | Process | chromatin organization | -1.386117403 | 0.772402 |
| 6 - PLANTS + FUNGI | Process | chromosome segregation | -1.279711176 | 1 |
| 6 - PLANTS + FUNGI | Process | cofactor metabolic process | -0.220817487 | 1 |
| 6 - PLANTS + FUNGI | Process | conjugation | -2.073260299 | 1 |
| 6 - PLANTS + FUNGI | Process | cytokinesis | -0.399292792 | 1 |
| 6 - PLANTS + FUNGI | Process | cytoplasmic translation | (none in phylogroup) | 0.05009132 |
| 6 - PLANTS + FUNGI | Process | cytoskeleton organization | -0.990798139 | 1 |
| 6 - PLANTS + FUNGI | Process | DNA recombination | -1.402567924 | 1 |
| 6 - PLANTS + FUNGI | Process | DNA repair | -1.463423129 | 0.6101195 |
| 6 - PLANTS + FUNGI | Process | DNA replication | (none in phylogroup) | 0.1580295 |
| 6 - PLANTS + FUNGI | Process | endocytosis | -0.78785808 | 1 |
| 6 - PLANTS + FUNGI | Process | endosome transport | -1.43171427 | 1 |
| 6 - PLANTS + FUNGI | Process | exocytosis | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | generation of precursor metabolites and energy | -1.569217793 | 0.977814 |
| 6 - PLANTS + FUNGI | Process | Golgi vesicle transport | -0.718595418 | 1 |
| 6 - PLANTS + FUNGI | Process | histone modification | (none in phylogroup) | 0.5864367 |
| 6 - PLANTS + FUNGI | Process | invasive growth in response to glucose limitation | -0.875320921 | 1 |
| 6 - PLANTS + FUNGI | Process | ion transport | 0.763681419 | 0.8283064 |
| 6 - PLANTS + FUNGI | Process | lipid metabolic process | 1.155998842 | 0.007215541 |
| 6 - PLANTS + FUNGI | Process | lipid transport | 1.989749499 | 0.01227145 |
| 6 - PLANTS + FUNGI | Process | meiotic cell cycle | -1.551623733 | 1 |
| 6 - PLANTS + FUNGI | Process | membrane fusion | -0.488297798 | 1 |
| 6 - PLANTS + FUNGI | Process | membrane invagination | 0.29460408 | 1 |
| 6 - PLANTS + FUNGI | Process | mitochondrial translation | -0.694748676 | 1 |
| 6 - PLANTS + FUNGI | Process | mitochondrion organization | -0.739836565 | 1 |
| 6 - PLANTS + FUNGI | Process | mitotic cell cycle | -1.030714604 | 0.908714 |
| 6 - PLANTS + FUNGI | Process | mRNA processing | -1.686711357 | 0.7504537 |
| 6 - PLANTS + FUNGI | Process | nuclear transport | -0.921713827 | 1 |
| 6 - PLANTS + FUNGI | Process | nucleobase, nucleoside and nucleotide metabolic process | 0.59547056 | 1 |
| 6 - PLANTS + FUNGI | Process | nucleobase, nucleoside, nucleotide and nucleic acid transport | 0.286142501 | 1 |
| 6 - PLANTS + FUNGI | Process | nucleus organization | 0.549176907 | 1 |
| 6 - PLANTS + FUNGI | Process | oligosaccharide metabolic process | -0.412348945 | 1 |
| 6 - PLANTS + FUNGI | Process | organelle assembly | -0.560447584 | 1 |
| 6 - PLANTS + FUNGI | Process | organelle fission | -1.202895579 | 1 |
| 6 - PLANTS + FUNGI | Process | organelle fusion | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | organelle inheritance | -0.903335297 | 1 |
| 6 - PLANTS + FUNGI | Process | peptidyl-amino acid modification | 0.549176907 | 1 |
| 6 - PLANTS + FUNGI | Process | peroxisome organization | 0.29460408 | 1 |
| 6 - PLANTS + FUNGI | Process | protein acylation | -1.180175503 | 1 |
| 6 - PLANTS + FUNGI | Process | protein alkylation | 0.475176326 | 1 |
| 6 - PLANTS + FUNGI | Process | protein complex biogenesis | -2.937605199 | 0.09578436 |
| 6 - PLANTS + FUNGI | Process | protein dephosphorylation | 1.134139408 | 0.8316331 |
| 6 - PLANTS + FUNGI | Process | protein folding | -1.662327198 | 1 |
| 6 - PLANTS + FUNGI | Process | protein glycosylation | 0.273542465 | 1 |
| 6 - PLANTS + FUNGI | Process | protein lipidation | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | protein maturation | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | protein modification by small protein conjugation or removal | -0.060876574 | 1 |
| 6 - PLANTS + FUNGI | Process | protein phosphorylation | 0.366054603 | 1 |
| 6 - PLANTS + FUNGI | Process | protein targeting | -0.640300892 | 1 |
| 6 - PLANTS + FUNGI | Process | proteolysis involved in cellular protein catabolic process | -0.187788687 | 1 |
| 6 - PLANTS + FUNGI | Process | pseudohyphal growth | -0.26898477 | 1 |
| 6 - PLANTS + FUNGI | Process | regulation of cell cycle | -1.164827538 | 1 |
| 6 - PLANTS + FUNGI | Process | regulation of DNA metabolic process | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | regulation of organelle organization | -2.236318581 | 0.8391343 |
| 6 - PLANTS + FUNGI | Process | regulation of protein modification process | 0.819824497 | 1 |
| 6 - PLANTS + FUNGI | Process | regulation of translation | -0.645839075 | 1 |
| 6 - PLANTS + FUNGI | Process | regulation of transport | 0.549176907 | 1 |
| 6 - PLANTS + FUNGI | Process | response to chemical stimulus | 0.298853553 | 1 |
| 6 - PLANTS + FUNGI | Process | response to DNA damage stimulus | -1.342446932 | 0.4810814 |
| 6 - PLANTS + FUNGI | Process | response to heat | -0.488297798 | 1 |
| 6 - PLANTS + FUNGI | Process | response to osmotic stress | 1.414634784 | 0.1252008 |
| 6 - PLANTS + FUNGI | Process | response to oxidative stress | 0.70964158 | 1 |
| 6 - PLANTS + FUNGI | Process | response to starvation | 0.56828573 | 1 |
| 6 - PLANTS + FUNGI | Process | ribosomal large subunit biogenesis | -1.560447584 | 1 |
| 6 - PLANTS + FUNGI | Process | ribosomal small subunit biogenesis | 0.427870611 | 1 |
| 6 - PLANTS + FUNGI | Process | ribosomal subunit export from nucleus | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | ribosome assembly | -1.085538629 | 1 |
| 6 - PLANTS + FUNGI | Process | RNA catabolic process | -1.944362566 | 1 |
| 6 - PLANTS + FUNGI | Process | RNA modification | -2.46968212 | 0.4850955 |
| 6 - PLANTS + FUNGI | Process | RNA splicing | -0.715965161 | 1 |
| 6 - PLANTS + FUNGI | Process | rRNA processing | -1.070792043 | 0.7983345 |
| 6 - PLANTS + FUNGI | Process | signaling | 0.121539371 | 1 |
| 6 - PLANTS + FUNGI | Process | snoRNA processing | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | sporulation | 0.393382235 | 1 |
| 6 - PLANTS + FUNGI | Process | telomere organization | -1.247289699 | 1 |
| 6 - PLANTS + FUNGI | Process | transcription elongation, DNA-dependent | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | transcription from RNA polymerase I promoter | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | transcription from RNA polymerase II promoter | -1.608887939 | 0.05071963 |
| 6 - PLANTS + FUNGI | Process | transcription from RNA polymerase III promoter | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | transcription initiation, DNA-dependent | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | transcription termination, DNA-dependent | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | translational elongation | -2.573582986 | 0.01467001 |
| 6 - PLANTS + FUNGI | Process | translational initiation | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | transmembrane transport | 0.942955287 | 0.192015 |
| 6 - PLANTS + FUNGI | Process | transposition | (none in phylogroup) | 0.40767 |
| 6 - PLANTS + FUNGI | Process | tRNA aminoacylation for protein translation | (none in phylogroup) | 1 |
| 6 - PLANTS + FUNGI | Process | tRNA processing | -0.232642923 | 1 |
| 6 - PLANTS + FUNGI | Process | vacuole organization | 0.096664703 | 1 |
| 6 - PLANTS + FUNGI | Process | vesicle organization | (none in phylogroup) | 0.971396 |
| 6 - PLANTS + FUNGI | Process | vitamin metabolic process | 1.564443664 | 0.1657067 |
| 7 - FUNGI | Component | cell cortex | 0.316817469 | 0.3157589 |
| 7 - FUNGI | Component | cell wall | 0.838783451 | 6.01E-07 |
| 7 - FUNGI | Component | cellular bud | 0.720179572 | 7.36E-09 |
| 7 - FUNGI | Component | cellular component unknown | 0.381695183 | 2.00E-08 |
| 7 - FUNGI | Component | chromosome | 0.332735766 | 0.006226334 |
| 7 - FUNGI | Component | cytoplasm | -0.203498105 | 4.14E-19 |
| 7 - FUNGI | Component | cytoplasmic membrane-bounded vesicle | -0.375527035 | 0.8339068 |
| 7 - FUNGI | Component | cytoskeleton | 0.437569628 | 0.002845123 |
| 7 - FUNGI | Component | endomembrane system | -0.089178259 | 1 |
| 7 - FUNGI | Component | endoplasmic reticulum | 0.091652037 | 1 |
| 7 - FUNGI | Component | extracellular region | 0.453511172 | 0.9389088 |
| 7 - FUNGI | Component | Golgi apparatus | -0.100462518 | 1 |
| 7 - FUNGI | Component | membrane | 0.094926592 | 0.3268965 |
| 7 - FUNGI | Component | membrane fraction | -0.077003545 | 1 |
| 7 - FUNGI | Component | microtubule organizing center | 0.488059398 | 0.1247801 |
| 7 - FUNGI | Component | mitochondrial envelope | -0.118030813 | 1 |
| 7 - FUNGI | Component | mitochondrion | -0.188648901 | 0.01599774 |
| 7 - FUNGI | Component | nucleolus | -1.132498657 | 1.21E-12 |
| 7 - FUNGI | Component | nucleus | 0.011681609 | 1 |
| 7 - FUNGI | Component | peroxisome | -0.426946016 | 1 |
| 7 - FUNGI | Component | plasma membrane | 0.431065356 | 0.000265231 |
| 7 - FUNGI | Component | ribosome | -0.666070444 | 3.46E-06 |
| 7 - FUNGI | Component | site of polarized growth | 0.576228232 | 2.00E-06 |
| 7 - FUNGI | Component | vacuole | -0.026154403 | 1 |
| 7 - FUNGI | Function | ATPase activity | -1.383334785 | 7.18E-12 |
| 7 - FUNGI | Function | cytoskeletal protein binding | 0.129447333 | 1 |
| 7 - FUNGI | Function | DNA binding | 0.589952328 | 2.42E-10 |
| 7 - FUNGI | Function | enzyme binding | 0.47048425 | 0.5702022 |
| 7 - FUNGI | Function | enzyme regulator activity | 0.481241617 | 0.00036813 |
| 7 - FUNGI | Function | GTPase activity | -0.62449134 | 0.615921 |
| 7 - FUNGI | Function | helicase activity | -0.170112949 | 1 |
| 7 - FUNGI | Function | histone binding | -0.120072267 | 1 |
| 7 - FUNGI | Function | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | -0.722338664 | 0.3148047 |
| 7 - FUNGI | Function | hydrolase activity, acting on glycosyl bonds | -0.587198277 | 0.971396 |
| 7 - FUNGI | Function | ion binding | -0.39016143 | 1 |
| 7 - FUNGI | Function | isomerase activity | -1.324931058 | 0.01222004 |
| 7 - FUNGI | Function | kinase activity | -0.658399431 | 0.001933161 |
| 7 - FUNGI | Function | ligase activity | -0.854611124 | 0.000136695 |
| 7 - FUNGI | Function | lipid binding | 0.194298477 | 1 |
| 7 - FUNGI | Function | lyase activity | -1.545152381 | 0.000101428 |
| 7 - FUNGI | Function | methyltransferase activity | -1.596377704 | 4.25E-05 |
| 7 - FUNGI | Function | molecular function unknown | 0.56875765 | 0 |
| 7 - FUNGI | Function | mRNA binding | -0.570043556 | 0.455073 |
| 7 - FUNGI | Function | nuclease activity | -0.759521355 | 0.009371203 |
| 7 - FUNGI | Function | nucleic acid binding transcription factor activity | 1.186030861 | 0 |
| 7 - FUNGI | Function | nucleotidyltransferase activity | -2.727257506 | 4.69E-12 |
| 7 - FUNGI | Function | oxidoreductase activity | -0.793210579 | 2.47E-06 |
| 7 - FUNGI | Function | peptidase activity | -1.197755704 | 1.20E-05 |
| 7 - FUNGI | Function | phosphatase activity | -0.448097697 | 0.6074449 |
| 7 - FUNGI | Function | protein binding transcription factor activity | 0.932456784 | 3.13E-07 |
| 7 - FUNGI | Function | protein binding, bridging | 0.186030861 | 1 |
| 7 - FUNGI | Function | protein transporter activity | -0.357111464 | 1 |
| 7 - FUNGI | Function | RNA binding | -2.465499257 | 2.16E-84 |
| 7 - FUNGI | Function | RNA modification guide activity | (none in phylogroup) | 6.20E-12 |
| 7 - FUNGI | Function | rRNA binding | -4.031199855 | 5.86E-13 |
| 7 - FUNGI | Function | signal transducer activity | 0.273493702 | 1 |
| 7 - FUNGI | Function | small conjugating protein binding | -0.217866081 | 1 |
| 7 - FUNGI | Function | structural constituent of ribosome | -0.900623521 | 3.56E-06 |
| 7 - FUNGI | Function | structural molecule activity | -0.12675658 | 1 |
| 7 - FUNGI | Function | transcription factor binding | 0.903887632 | 0.001474174 |
| 7 - FUNGI | Function | transferase activity, transferring acyl groups | -0.255001847 | 1 |
| 7 - FUNGI | Function | transferase activity, transferring alkyl or aryl (other than methyl) groups | -1.455515168 | 0.05114955 |
| 7 - FUNGI | Function | transferase activity, transferring glycosyl groups | -0.048434393 | 1 |
| 7 - FUNGI | Function | translation factor activity, nucleic acid binding | -4.015603 | 4.75E-06 |
| 7 - FUNGI | Function | transmembrane transporter activity | -0.219668774 | 0.661148 |
| 7 - FUNGI | Function | triplet codon-amino acid adaptor activity | (none in phylogroup) | 3.02E-56 |
| 7 - FUNGI | Function | unfolded protein binding | -0.23620214 | 1 |
| 7 - FUNGI | Process | amino acid transport | 0.633489838 | 0.07736126 |
| 7 - FUNGI | Process | biological process unknown | 0.573019031 | 0 |
| 7 - FUNGI | Process | carbohydrate metabolic process | -0.215033696 | 0.8423998 |
| 7 - FUNGI | Process | carbohydrate transport | -0.418040463 | 1 |
| 7 - FUNGI | Process | cell budding | 0.268493021 | 1 |
| 7 - FUNGI | Process | cell morphogenesis | 0.468430592 | 0.615921 |
| 7 - FUNGI | Process | cell wall organization or biogenesis | 0.675686402 | 4.45E-09 |
| 7 - FUNGI | Process | cellular amino acid metabolic process | -1.35453752 | 4.69E-12 |
| 7 - FUNGI | Process | cellular ion homeostasis | -0.077003545 | 1 |
| 7 - FUNGI | Process | cellular respiration | -0.37174681 | 1 |
| 7 - FUNGI | Process | chromatin organization | 0.166039386 | 1 |
| 7 - FUNGI | Process | chromosome segregation | 0.361880696 | 0.1553753 |
| 7 - FUNGI | Process | cofactor metabolic process | -2.372459428 | 1.73E-15 |
| 7 - FUNGI | Process | conjugation | 0.544484832 | 0.004918678 |
| 7 - FUNGI | Process | cytokinesis | 0.533954164 | 0.009280359 |
| 7 - FUNGI | Process | cytoplasmic translation | -3.350022039 | 1.51E-22 |
| 7 - FUNGI | Process | cytoskeleton organization | 0.211909493 | 0.6643103 |
| 7 - FUNGI | Process | DNA recombination | 0.285566535 | 0.4465299 |
| 7 - FUNGI | Process | DNA repair | -0.337531095 | 0.238878 |
| 7 - FUNGI | Process | DNA replication | -0.315163282 | 0.8825475 |
| 7 - FUNGI | Process | endocytosis | 0.132449821 | 1 |
| 7 - FUNGI | Process | endosome transport | -0.135897234 | 1 |
| 7 - FUNGI | Process | exocytosis | -0.346190178 | 1 |
| 7 - FUNGI | Process | generation of precursor metabolites and energy | -0.273400758 | 0.9535202 |
| 7 - FUNGI | Process | Golgi vesicle transport | -0.393031038 | 0.2211471 |
| 7 - FUNGI | Process | histone modification | -0.032609426 | 1 |
| 7 - FUNGI | Process | invasive growth in response to glucose limitation | 0.294965233 | 1 |
| 7 - FUNGI | Process | ion transport | -0.099199291 | 1 |
| 7 - FUNGI | Process | lipid metabolic process | -0.571267735 | 0.001447834 |
| 7 - FUNGI | Process | lipid transport | -0.299395966 | 1 |
| 7 - FUNGI | Process | meiotic cell cycle | 0.308977921 | 0.2415658 |
| 7 - FUNGI | Process | membrane fusion | -0.192480762 | 1 |
| 7 - FUNGI | Process | membrane invagination | 0.120935833 | 1 |
| 7 - FUNGI | Process | mitochondrial translation | 0.264033373 | 0.6743476 |
| 7 - FUNGI | Process | mitochondrion organization | 0.174890303 | 0.6777927 |
| 7 - FUNGI | Process | mitotic cell cycle | 0.33549176 | 0.01577573 |
| 7 - FUNGI | Process | mRNA processing | -0.484003725 | 0.06998844 |
| 7 - FUNGI | Process | nuclear transport | -0.473893697 | 0.1316088 |
| 7 - FUNGI | Process | nucleobase, nucleoside and nucleotide metabolic process | -0.886319983 | 5.97E-05 |
| 7 - FUNGI | Process | nucleobase, nucleoside, nucleotide and nucleic acid transport | -0.115477693 | 1 |
| 7 - FUNGI | Process | nucleus organization | 0.318925131 | 1 |
| 7 - FUNGI | Process | oligosaccharide metabolic process | -0.70149441 | 0.9465203 |
| 7 - FUNGI | Process | organelle assembly | -1.679668047 | 3.68E-05 |
| 7 - FUNGI | Process | organelle fission | 0.315313878 | 0.3764599 |
| 7 - FUNGI | Process | organelle fusion | -0.214507069 | 1 |
| 7 - FUNGI | Process | organelle inheritance | -0.105017921 | 1 |
| 7 - FUNGI | Process | peptidyl-amino acid modification | -0.347651135 | 1 |
| 7 - FUNGI | Process | peroxisome organization | 0.507958956 | 0.1154578 |
| 7 - FUNGI | Process | protein acylation | -0.221393454 | 1 |
| 7 - FUNGI | Process | protein alkylation | -0.813969139 | 0.5813516 |
| 7 - FUNGI | Process | protein complex biogenesis | 0.148288767 | 1 |
| 7 - FUNGI | Process | protein dephosphorylation | -0.039528839 | 1 |
| 7 - FUNGI | Process | protein folding | -0.629544568 | 0.1998637 |
| 7 - FUNGI | Process | protein glycosylation | 0.099874217 | 1 |
| 7 - FUNGI | Process | protein lipidation | -1.110950877 | 0.1412847 |
| 7 - FUNGI | Process | protein maturation | -0.455515168 | 1 |
| 7 - FUNGI | Process | protein modification by small protein conjugation or removal | -0.180097038 | 1 |
| 7 - FUNGI | Process | protein phosphorylation | -0.271014165 | 1 |
| 7 - FUNGI | Process | protein targeting | -0.122091434 | 1 |
| 7 - FUNGI | Process | proteolysis involved in cellular protein catabolic process | -0.203915657 | 1 |
| 7 - FUNGI | Process | pseudohyphal growth | 0.249224687 | 1 |
| 7 - FUNGI | Process | regulation of cell cycle | 0.227851037 | 0.6741498 |
| 7 - FUNGI | Process | regulation of DNA metabolic process | 0.013194264 | 1 |
| 7 - FUNGI | Process | regulation of organelle organization | 0.118392144 | 1 |
| 7 - FUNGI | Process | regulation of protein modification process | 0.115641533 | 1 |
| 7 - FUNGI | Process | regulation of translation | -0.127629618 | 1 |
| 7 - FUNGI | Process | regulation of transport | -0.077003545 | 1 |
| 7 - FUNGI | Process | response to chemical stimulus | 0.312270858 | 0.01398745 |
| 7 - FUNGI | Process | response to DNA damage stimulus | -0.205327642 | 0.9063606 |
| 7 - FUNGI | Process | response to heat | 0.681988356 | 0.06125594 |
| 7 - FUNGI | Process | response to osmotic stress | 0.272330707 | 1 |
| 7 - FUNGI | Process | response to oxidative stress | -0.509114558 | 0.5280079 |
| 7 - FUNGI | Process | response to starvation | -0.020420016 | 1 |
| 7 - FUNGI | Process | ribosomal large subunit biogenesis | -1.264630548 | 0.001331447 |
| 7 - FUNGI | Process | ribosomal small subunit biogenesis | -1.986805736 | 1.08E-09 |
| 7 - FUNGI | Process | ribosomal subunit export from nucleus | -0.884358467 | 0.3931591 |
| 7 - FUNGI | Process | ribosome assembly | -2.789721593 | 3.67E-06 |
| 7 - FUNGI | Process | RNA catabolic process | -0.326617435 | 1 |
| 7 - FUNGI | Process | RNA modification | -3.758827585 | 2.15E-21 |
| 7 - FUNGI | Process | RNA splicing | -0.502610286 | 0.1525779 |
| 7 - FUNGI | Process | rRNA processing | -1.926978101 | 2.98E-23 |
| 7 - FUNGI | Process | signaling | 0.443828618 | 0.001138231 |
| 7 - FUNGI | Process | snoRNA processing | -0.579503885 | 1 |
| 7 - FUNGI | Process | sporulation | 0.274161771 | 0.6545212 |
| 7 - FUNGI | Process | telomere organization | 0.048527337 | 1 |
| 7 - FUNGI | Process | transcription elongation, DNA-dependent | 0.022532129 | 1 |
| 7 - FUNGI | Process | transcription from RNA polymerase I promoter | -0.237468217 | 1 |
| 7 - FUNGI | Process | transcription from RNA polymerase II promoter | 0.50113769 | 7.50E-09 |
| 7 - FUNGI | Process | transcription from RNA polymerase III promoter | -0.813969139 | 0.5813516 |
| 7 - FUNGI | Process | transcription initiation, DNA-dependent | 0.19601495 | 1 |
| 7 - FUNGI | Process | transcription termination, DNA-dependent | -1.39893164 | 0.1814844 |
| 7 - FUNGI | Process | translational elongation | -5.27776595 | 1.46E-56 |
| 7 - FUNGI | Process | translational initiation | -1.129470965 | 0.04231877 |
| 7 - FUNGI | Process | transmembrane transport | 0.061894561 | 1 |
| 7 - FUNGI | Process | transposition | -2.246928546 | 1.44E-09 |
| 7 - FUNGI | Process | tRNA aminoacylation for protein translation | -2.70149441 | 0.00118566 |
| 7 - FUNGI | Process | tRNA processing | -1.299395966 | 0.000172219 |
| 7 - FUNGI | Process | vacuole organization | -0.054977239 | 1 |
| 7 - FUNGI | Process | vesicle organization | -0.43689949 | 0.9347144 |
| 7 - FUNGI | Process | vitamin metabolic process | -1.724701801 | 0.005533816 |
| 8 - MINOR CLUSTERS | Component | cell cortex | 0.066189206 | 1 |
| 8 - MINOR CLUSTERS | Component | cell wall | -0.757619648 | 1 |
| 8 - MINOR CLUSTERS | Component | cellular bud | 0.648521451 | 0.833180878 |
| 8 - MINOR CLUSTERS | Component | cellular component unknown | -0.182090964 | 1 |
| 8 - MINOR CLUSTERS | Component | chromosome | -1.571334734 | 0.21599174 |
| 8 - MINOR CLUSTERS | Component | cytoplasm | 0.155414156 | 0.3289849 |
| 8 - MINOR CLUSTERS | Component | cytoplasmic membrane-bounded vesicle | -1.899173792 | 1 |
| 8 - MINOR CLUSTERS | Component | cytoskeleton | -0.550024228 | 1 |
| 8 - MINOR CLUSTERS | Component | endomembrane system | -1.315558975 | 0.322558417 |
| 8 - MINOR CLUSTERS | Component | endoplasmic reticulum | -0.916405922 | 0.780397906 |
| 8 - MINOR CLUSTERS | Component | extracellular region | 0.087405692 | 1 |
| 8 - MINOR CLUSTERS | Component | Golgi apparatus | -2.696865617 | 0.290188003 |
| 8 - MINOR CLUSTERS | Component | membrane | 0.25933583 | 0.795612038 |
| 8 - MINOR CLUSTERS | Component | membrane fraction | 0.470734332 | 1 |
| 8 - MINOR CLUSTERS | Component | microtubule organizing center | -1.347531364 | 1 |
| 8 - MINOR CLUSTERS | Component | mitochondrial envelope | 0.100826208 | 1 |
| 8 - MINOR CLUSTERS | Component | mitochondrion | 0.747135961 | 0.000302133 |
| 8 - MINOR CLUSTERS | Component | nucleolus | -1.470589761 | 0.321796128 |
| 8 - MINOR CLUSTERS | Component | nucleus | -0.509035817 | 0.042063129 |
| 8 - MINOR CLUSTERS | Component | peroxisome | -0.180074619 | 1 |
| 8 - MINOR CLUSTERS | Component | plasma membrane | 1.252330013 | 0.001507919 |
| 8 - MINOR CLUSTERS | Component | ribosome | -1.617138424 | 0.17857877 |
| 8 - MINOR CLUSTERS | Component | site of polarized growth | 0.015744707 | 1 |
| 8 - MINOR CLUSTERS | Component | vacuole | 1.128856136 | 0.040416221 |
| 8 - MINOR CLUSTERS | Function | ATPase activity | -1.418234356 | 0.71696558 |
| 8 - MINOR CLUSTERS | Function | cytoskeletal protein binding | -0.180074619 | 1 |
| 8 - MINOR CLUSTERS | Function | DNA binding | -1.57555932 | 0.21599174 |
| 8 - MINOR CLUSTERS | Function | enzyme binding | 0.556890976 | 1 |
| 8 - MINOR CLUSTERS | Function | enzyme regulator activity | -0.314211291 | 1 |
| 8 - MINOR CLUSTERS | Function | GTPase activity | -0.99059682 | 1 |
| 8 - MINOR CLUSTERS | Function | helicase activity | (none in phylogroup) | 0.838537397 |
| 8 - MINOR CLUSTERS | Function | histone binding | 0.754830353 | 1 |
| 8 - MINOR CLUSTERS | Function | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 0.911555857 | 0.859008811 |
| 8 - MINOR CLUSTERS | Function | hydrolase activity, acting on glycosyl bonds | 0.872666844 | 0.975916755 |
| 8 - MINOR CLUSTERS | Function | ion binding | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Function | isomerase activity | 1.00940318 | 0.795612038 |
| 8 - MINOR CLUSTERS | Function | kinase activity | -0.493990193 | 1 |
| 8 - MINOR CLUSTERS | Function | ligase activity | 0.342447307 | 1 |
| 8 - MINOR CLUSTERS | Function | lipid binding | 0.399349699 | 1 |
| 8 - MINOR CLUSTERS | Function | lyase activity | 2.274608685 | 2.00E-05 |
| 8 - MINOR CLUSTERS | Function | methyltransferase activity | -1.58397156 | 1 |
| 8 - MINOR CLUSTERS | Function | molecular function unknown | -0.421607955 | 0.17857877 |
| 8 - MINOR CLUSTERS | Function | mRNA binding | 0.179328182 | 1 |
| 8 - MINOR CLUSTERS | Function | nuclease activity | -0.595112118 | 1 |
| 8 - MINOR CLUSTERS | Function | nucleic acid binding transcription factor activity | -0.617138424 | 1 |
| 8 - MINOR CLUSTERS | Function | nucleotidyltransferase activity | -1.977885768 | 1 |
| 8 - MINOR CLUSTERS | Function | oxidoreductase activity | 1.943222103 | 1.06E-10 |
| 8 - MINOR CLUSTERS | Function | peptidase activity | -0.670776388 | 1 |
| 8 - MINOR CLUSTERS | Function | phosphatase activity | 0.827342853 | 0.822526449 |
| 8 - MINOR CLUSTERS | Function | protein binding transcription factor activity | -0.386525496 | 1 |
| 8 - MINOR CLUSTERS | Function | protein binding, bridging | -0.649559902 | 1 |
| 8 - MINOR CLUSTERS | Function | protein transporter activity | 0.169867853 | 1 |
| 8 - MINOR CLUSTERS | Function | RNA binding | -2.131165018 | 7.86E-05 |
| 8 - MINOR CLUSTERS | Function | RNA modification guide activity | (none in phylogroup) | 0.975916755 |
| 8 - MINOR CLUSTERS | Function | rRNA binding | -1.696865617 | 1 |
| 8 - MINOR CLUSTERS | Function | signal transducer activity | 0.5203651 | 1 |
| 8 - MINOR CLUSTERS | Function | small conjugating protein binding | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Function | structural constituent of ribosome | -1.958606705 | 0.331314205 |
| 8 - MINOR CLUSTERS | Function | structural molecule activity | -1.625312356 | 0.17857877 |
| 8 - MINOR CLUSTERS | Function | transcription factor binding | 1.034938272 | 0.838537397 |
| 8 - MINOR CLUSTERS | Function | transferase activity, transferring acyl groups | -0.380099227 | 1 |
| 8 - MINOR CLUSTERS | Function | transferase activity, transferring alkyl or aryl (other than methyl) groups | 1.878819071 | 0.094496548 |
| 8 - MINOR CLUSTERS | Function | transferase activity, transferring glycosyl groups | 0.5203651 | 1 |
| 8 - MINOR CLUSTERS | Function | translation factor activity, nucleic acid binding | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Function | transmembrane transporter activity | 1.529702964 | 4.28E-06 |
| 8 - MINOR CLUSTERS | Function | triplet codon-amino acid adaptor activity | (none in phylogroup) | 0.001506699 |
| 8 - MINOR CLUSTERS | Function | unfolded protein binding | -0.223795996 | 1 |
| 8 - MINOR CLUSTERS | Process | amino acid transport | 2.382861576 | 0.001366843 |
| 8 - MINOR CLUSTERS | Process | biological process unknown | -0.010942089 | 1 |
| 8 - MINOR CLUSTERS | Process | carbohydrate metabolic process | 0.640659333 | 0.653834891 |
| 8 - MINOR CLUSTERS | Process | carbohydrate transport | -0.405634319 | 1 |
| 8 - MINOR CLUSTERS | Process | cell budding | 0.754830353 | 0.955694775 |
| 8 - MINOR CLUSTERS | Process | cell morphogenesis | 1.257330694 | 0.701295851 |
| 8 - MINOR CLUSTERS | Process | cell wall organization or biogenesis | 0.237431136 | 1 |
| 8 - MINOR CLUSTERS | Process | cellular amino acid metabolic process | 1.742757521 | 1.06E-06 |
| 8 - MINOR CLUSTERS | Process | cellular ion homeostasis | 0.707133611 | 0.877272032 |
| 8 - MINOR CLUSTERS | Process | cellular respiration | -0.096306261 | 1 |
| 8 - MINOR CLUSTERS | Process | chromatin organization | -1.347531364 | 0.795612038 |
| 8 - MINOR CLUSTERS | Process | chromosome segregation | -2.234522403 | 0.795612038 |
| 8 - MINOR CLUSTERS | Process | cofactor metabolic process | 1.672368193 | 0.000308741 |
| 8 - MINOR CLUSTERS | Process | conjugation | 0.791667121 | 0.795612038 |
| 8 - MINOR CLUSTERS | Process | cytokinesis | -0.977885768 | 1 |
| 8 - MINOR CLUSTERS | Process | cytoplasmic translation | (none in phylogroup) | 0.090789252 |
| 8 - MINOR CLUSTERS | Process | cytoskeleton organization | -0.138254443 | 1 |
| 8 - MINOR CLUSTERS | Process | DNA recombination | -2.35737915 | 0.679617705 |
| 8 - MINOR CLUSTERS | Process | DNA repair | -2.003196856 | 0.321796128 |
| 8 - MINOR CLUSTERS | Process | DNA replication | (none in phylogroup) | 0.232105254 |
| 8 - MINOR CLUSTERS | Process | endocytosis | -0.157706805 | 1 |
| 8 - MINOR CLUSTERS | Process | endosome transport | (none in phylogroup) | 0.908761713 |
| 8 - MINOR CLUSTERS | Process | exocytosis | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | generation of precursor metabolites and energy | 1.382861576 | 0.011777439 |
| 8 - MINOR CLUSTERS | Process | Golgi vesicle transport | (none in phylogroup) | 0.068789925 |
| 8 - MINOR CLUSTERS | Process | histone modification | -1.742669306 | 1 |
| 8 - MINOR CLUSTERS | Process | invasive growth in response to glucose limitation | 0.11437274 | 1 |
| 8 - MINOR CLUSTERS | Process | ion transport | 2.130798288 | 1.06E-06 |
| 8 - MINOR CLUSTERS | Process | lipid metabolic process | 0.11982717 | 1 |
| 8 - MINOR CLUSTERS | Process | lipid transport | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | meiotic cell cycle | -1.50643496 | 0.921383988 |
| 8 - MINOR CLUSTERS | Process | membrane fusion | (none in phylogroup) | 0.863915217 |
| 8 - MINOR CLUSTERS | Process | membrane invagination | -0.660207146 | 1 |
| 8 - MINOR CLUSTERS | Process | mitochondrial translation | -0.649559902 | 1 |
| 8 - MINOR CLUSTERS | Process | mitochondrion organization | -0.502002713 | 1 |
| 8 - MINOR CLUSTERS | Process | mitotic cell cycle | -1.717402548 | 0.284041948 |
| 8 - MINOR CLUSTERS | Process | mRNA processing | -2.641522583 | 0.322558417 |
| 8 - MINOR CLUSTERS | Process | nuclear transport | -1.461487554 | 0.981286918 |
| 8 - MINOR CLUSTERS | Process | nucleobase, nucleoside and nucleotide metabolic process | 1.318731238 | 0.012536372 |
| 8 - MINOR CLUSTERS | Process | nucleobase, nucleoside, nucleotide and nucleic acid transport | -1.99059682 | 1 |
| 8 - MINOR CLUSTERS | Process | nucleus organization | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | oligosaccharide metabolic process | 0.632839829 | 1 |
| 8 - MINOR CLUSTERS | Process | organelle assembly | (none in phylogroup) | 0.833180878 |
| 8 - MINOR CLUSTERS | Process | organelle fission | -2.157706805 | 0.838537397 |
| 8 - MINOR CLUSTERS | Process | organelle fusion | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | organelle inheritance | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | peptidyl-amino acid modification | 0.179328182 | 1 |
| 8 - MINOR CLUSTERS | Process | peroxisome organization | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | protein acylation | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | protein alkylation | 0.5203651 | 1 |
| 8 - MINOR CLUSTERS | Process | protein complex biogenesis | -1.307453925 | 0.838537397 |
| 8 - MINOR CLUSTERS | Process | protein dephosphorylation | 0.556890976 | 1 |
| 8 - MINOR CLUSTERS | Process | protein folding | -1.617138424 | 1 |
| 8 - MINOR CLUSTERS | Process | protein glycosylation | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | protein lipidation | -0.58397156 | 1 |
| 8 - MINOR CLUSTERS | Process | protein maturation | -0.443109024 | 1 |
| 8 - MINOR CLUSTERS | Process | protein modification by small protein conjugation or removal | -2.337615895 | 0.701295851 |
| 8 - MINOR CLUSTERS | Process | protein phosphorylation | 0.188850956 | 1 |
| 8 - MINOR CLUSTERS | Process | protein targeting | -1.180074619 | 0.795612038 |
| 8 - MINOR CLUSTERS | Process | proteolysis involved in cellular protein catabolic process | -2.727562414 | 0.272578313 |
| 8 - MINOR CLUSTERS | Process | pseudohyphal growth | 0.754830353 | 1 |
| 8 - MINOR CLUSTERS | Process | regulation of cell cycle | -0.374937522 | 1 |
| 8 - MINOR CLUSTERS | Process | regulation of DNA metabolic process | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | regulation of organelle organization | -1.191129807 | 1 |
| 8 - MINOR CLUSTERS | Process | regulation of protein modification process | 0.842293195 | 0.921383988 |
| 8 - MINOR CLUSTERS | Process | regulation of translation | (none in phylogroup) | 0.795612038 |
| 8 - MINOR CLUSTERS | Process | regulation of transport | 0.00940318 | 1 |
| 8 - MINOR CLUSTERS | Process | response to chemical stimulus | 0.693615927 | 0.3289849 |
| 8 - MINOR CLUSTERS | Process | response to DNA damage stimulus | -2.297258158 | 0.105407464 |
| 8 - MINOR CLUSTERS | Process | response to heat | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | response to osmotic stress | 0.652468636 | 1 |
| 8 - MINOR CLUSTERS | Process | response to oxidative stress | 0.737956535 | 0.975916755 |
| 8 - MINOR CLUSTERS | Process | response to starvation | -0.386525496 | 1 |
| 8 - MINOR CLUSTERS | Process | ribosomal large subunit biogenesis | -1.51525881 | 1 |
| 8 - MINOR CLUSTERS | Process | ribosomal small subunit biogenesis | -2.111903116 | 0.877272032 |
| 8 - MINOR CLUSTERS | Process | ribosomal subunit export from nucleus | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | ribosome assembly | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | RNA catabolic process | (none in phylogroup) | 0.480730399 |
| 8 - MINOR CLUSTERS | Process | RNA modification | (none in phylogroup) | 0.16363502 |
| 8 - MINOR CLUSTERS | Process | RNA splicing | -2.255738888 | 0.795612038 |
| 8 - MINOR CLUSTERS | Process | rRNA processing | (none in phylogroup) | 0.001672096 |
| 8 - MINOR CLUSTERS | Process | signaling | 0.166728145 | 1 |
| 8 - MINOR CLUSTERS | Process | snoRNA processing | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | sporulation | 0.438571008 | 1 |
| 8 - MINOR CLUSTERS | Process | telomere organization | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | transcription elongation, DNA-dependent | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | transcription from RNA polymerase I promoter | 0.594365681 | 1 |
| 8 - MINOR CLUSTERS | Process | transcription from RNA polymerase II promoter | -1.071451072 | 0.388650056 |
| 8 - MINOR CLUSTERS | Process | transcription from RNA polymerase III promoter | 1.1053276 | 0.795612038 |
| 8 - MINOR CLUSTERS | Process | transcription initiation, DNA-dependent | -1.386525496 | 1 |
| 8 - MINOR CLUSTERS | Process | transcription termination, DNA-dependent | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | translational elongation | (none in phylogroup) | 0.000546218 |
| 8 - MINOR CLUSTERS | Process | translational initiation | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | transmembrane transport | 0.888096884 | 0.328092341 |
| 8 - MINOR CLUSTERS | Process | transposition | (none in phylogroup) | 0.474723302 |
| 8 - MINOR CLUSTERS | Process | tRNA aminoacylation for protein translation | -0.367160171 | 1 |
| 8 - MINOR CLUSTERS | Process | tRNA processing | (none in phylogroup) | 0.65001518 |
| 8 - MINOR CLUSTERS | Process | vacuole organization | (none in phylogroup) | 1 |
| 8 - MINOR CLUSTERS | Process | vesicle organization | (none in phylogroup) | 0.877272032 |
| 8 - MINOR CLUSTERS | Process | vitamin metabolic process | 2.747135961 | 8.07E-06 |